Volume 10, Number 12—December 2004
Dispatch
Novel Avian Influenza H7N3 Strain Outbreak, British Columbia
Figure 1
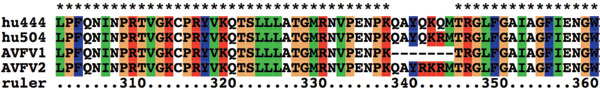
Figure 1. Alignment of the hemagglutinin cleavage region from four isolates of Fraser Valley H7N3 virus. A/Chicken/Canada/AVFV1/04 is designated AVFV1; A/Chicken/Canada/AVFV2/04 is designated AVFV2; A/Canada/444/04 (human) is Hu444, and A/Canada/504/04 (human) is Hu504). A 7–amino-acid (aa) insertion associated with the AVFV2 isolate and both human isolates is shown at aa 338.
Page created: October 26, 2024
Page updated: October 26, 2024
Page reviewed: October 26, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.