Volume 11, Number 11—November 2005
Letter
Ehrlichia ruminantium, Sudan
To the Editor: Ehrlichia ruminantium, the causative agent of heartwater, is transmitted by Amblyomma spp. ticks. Amblyomma variegatum ticks, which are found in the Caribbean and sub-Saharan Africa, except in certain areas of southern Africa, are major vectors of E. ruminantium (1–3). A. lepidum is also an important vector of heartwater, especially in eastern Sudan (4). However, few epidemiologic data exist on infection rates of Amblyomma spp. ticks and distribution of E. ruminantium in Sudan. A polymerase chain reaction (PCR) assay that uses DNA probe pCS20 has been developed for detecting E. ruminantium (5). Another PCR assay for the major antigen protein 1 gene (map1) has been used to differentiate strains of E. ruminantium (6,7). These PCR assays have high sensitivity and specificity for the amplification of E. ruminantium DNA (6,8). For epidemiologic study of E. ruminantium in Sudan, we used PCR to detect E. ruminantium DNA in ticks. We also sequenced PCR products to identify the genotype of E. ruminantium.
The pCS20 DNA fragment of E. ruminantium was detected in 8 (8.2%) of 97 A. variegatum ticks and 2 (1.9%) of 106 A. lepidum ticks (χ2= 3.123, by Yates correction). The nucleotide sequences (279 bp) obtained from 5 A. variegatum ticks and 1 A. lepidum tick were identical (GenBank accession no. AB218277). The sequences were similar to those of Welgevonden, Vosloo, and Ball3 strains from southern Africa and Gardel strain from the Caribbean islands (similarity = 99.64%). The pCS20 sequences obtained in this study were different from those of strains from western Africa.
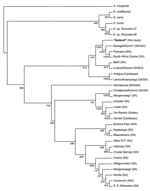
An 855-bp map1 nucleotide sequence obtained from 1 A. lepidum tick was provisionally named Gedaref (GenBank accession no. AB218278). The nucleotide sequence of Gedaref was found to be closely related to those of Senegal and Pokoase strains from western Africa and to South Africa Canine and Kümm1 strains from southern Africa (similarity = 90.53%–97.43%). Gedaref clustered with these 4 strains and with 6 other strains, including Kiswani from eastern Africa and Antigua from the Caribbean islands (Figure). In contrast, the nucleotide sequence of Gedaref showed 84.8% similarity with that of Um Banein, which has been known as the only strain of E. ruminantium in Sudan. Um Banein formed another cluster with Gardel, Lutale, and Umpala strains from southern Africa (Figure). The map1 coding sequence of Gedaref was closely related to those of strains Senegal, Ball3, South Africa Canine, and Pokoase (similarity = 92.61%–97.97%). Gedaref and these 4 strains formed a cluster and branch with Um Banein (similarity = 87.6%).
The novel E. ruminantium genotype Gedaref was detected in A. lepidum by PCR assays. This work has shown that another strain of E. ruminantium, in addition to the Um Banein strain, is present in Sudan. Since the Um Banein strain of E. ruminantium was isolated from sheep in 1984 (4), no other strain had been reported in Sudan. The map1 sequence of Gedaref has diverged from that of Um Banein. Gedaref formed a cluster with several strains that originated in southern and western Africa. As previously reported, the variation of map1 sequences of E. ruminantium strains does not reflect the geographic distribution of the strains (6). However, Gedaref was distinctively differentiated from the Um Banein strain. Gedaref was closely related to 3 southern African strains and a Caribbean strain in the pCS20 sequence but different from western African strains. The pCS20 sequence has been highly conserved among strains from western Africa (10), and the distribution of A. lepidum is limited to eastern Africa (3). If one considers the distribution of A. lepidum, results of genetic analyses in the pCS20 gene regions of Gedaref are important for epidemiologic research on E. ruminantium.
We detected pCS20 DNA specific for E. ruminantium in A. variegatum. This tick is widely distributed in Africa and is the most efficient vector of heartwater (3). Nevertheless, detection of E. ruminantium in A. variegatum in Sudan had not previously been reported. Our results show that A. variegatum is also an important vector of E. ruminantium in Sudan. A. variegatum ticks are also found North and South America, Southeast Asia, and Australia (3). Thus, the potential spread of E. ruminantium to livestock is a continuous threat in these regions from the importation of tick-infested and subclinically infected wild animals from Africa and the importation of livestock from Caribbean islands (1,2). We believe that A. variegatum plays an important role in the spread of heartwater because of its ability to adapt to various environments.
Acknowledgments
This article has been published with permission of the director general of the Animal Resources Research Corporation, Khartoum, Sudan.
This work was supported by Gakujyutsu-frontier cooperative research at Rakuno Gakuen University; the High-Tech Research Center at Rakuno Gakuen University; a grant-in-aid for cooperative research from Rakuno Gakuen University (2003); and a grant-in-aid for scientific research (C) (14560272) from the Ministry of Education, Science, Sports, and Culture of Japan.
References
- Uilenberg G. Heartwater (Cowdria ruminantium infection): current status. Adv Vet Sci Comp Med. 1983;27:427–80.PubMedGoogle Scholar
- Burridge MJ, Simmons LA, Peter TF, Mahan SM. Increasing risks of introduction of heartwater onto the American mainland associated with animal movements. Ann N Y Acad Sci. 2002;969:269–74. DOIPubMedGoogle Scholar
- Walker JB, Olwage A. The tick vectors of Cowdria ruminantium (Ixodoidea, Ixodidae, genus Amblyomma) and their distribution. Onderstepoort J Vet Res. 1987;54:353–79.PubMedGoogle Scholar
- Jongejan F, Morzaria SP, Shariff OA, Abdalla HM. Isolation and transmission of Cowdria ruminantium (causal agent of heartwater disease) in Blue Nile Province, Sudan. Vet Res Commun. 1984;8:141–5. DOIPubMedGoogle Scholar
- Peter TF, Deem SL, Barbet AF, Norval RAI, Simbi BH, Kelly PJ, Development and evaluation of PCR assay for detection of low levels of Cowdria ruminantium infection in Amblyomma ticks not detected by DNA probe. J Clin Microbiol. 1995;33:166–72.PubMedGoogle Scholar
- Allsopp MTEP, Dorfling CM, Maillard JC, Bensaid A, Haydon DT, van Heerden H, Ehrlichia ruminantium major antigenic protein gene (map1) variants are not geographically constrained and show no evidence of having evolved under positive selection pressure. J Clin Microbiol. 2001;39:4200–3. DOIPubMedGoogle Scholar
- Reddy GR, Sulsona CR, Harrison RH, Mahan SM, Burridge MJ, Barbet AF. Sequence heterogeneity of the major antigenic protein 1 genes from Cowdria ruminantium isolates from different geographical areas. Clin Diagn Lab Immunol. 1996;3:417–22.PubMedGoogle Scholar
- Peter TF, Barbet AF, Alleman AR, Simbi BH, Burridge MJ, Mahan SM. Detection of the agent of heartwater, Cowdria ruminantium, in Amblyomma ticks by PCR: validation and application of the assay to field ticks. J Clin Microbiol. 2000;38:1539–44.PubMedGoogle Scholar
- Morita C, Hussein ARME, Matsuda E, Gabbar KMAA, Muramatsu Y, Rahman MBA, Spotted fever group rickettsiae from ticks captured in Sudan. Jpn J Infect Dis. 2004;57:107–9.PubMedGoogle Scholar
- Allsopp MTEP, van Heerden H, Steyn HC, Allsopp BA. Phylogenetic relationships among Ehrlichia ruminantium isolates. Ann N Y Acad Sci. 2003;990:685–91. DOIPubMedGoogle Scholar
Figure
Cite This ArticleRelated Links
Table of Contents – Volume 11, Number 11—November 2005
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Yasukazu Muramatsu, Department of Veterinary Public Health, School of Veterinary Medicine, Rakuno Gakuen University, 582-1, Bunkyodai-Midorimachi, Ebetsu, Hokkaido, 069-8501, Japan; fax: 81-011-387-5890
Top