Volume 12, Number 3—March 2006
Research
Host Feeding Patterns of Culex Mosquitoes and West Nile Virus Transmission, Northeastern United States
Cite This Article
Citation for Media
Abstract
To evaluate the role of Culex mosquitoes as enzootic and epidemic vectors for WNV, we identified the source of vertebrate blood by polymerase chain reaction amplification and sequencing portions of the cytochrome b gene of mitochondrial DNA. All Cx. restuans and 93% of Cx. pipiens acquired blood from avian hosts; Cx. salinarius fed frequently on both mammals (53%) and birds (36%). Mixed-blood meals were detected in 11% and 4% of Cx. salinarius and Cx. pipiens, respectively. American robin was the most common source of vertebrate blood for Cx. pipiens (38%) and Cx. restuans (37%). American crow represented <1% of the blood meals in Cx. pipiens and none in Cx. restuans. Human-derived blood meals were identified from 2 Cx. salinarius and 1 Cx. pipiens. Results suggest that Cx. salinarius is an important bridge vector to humans, while Cx. pipiens and Cx. restuans are more efficient enzootic vectors in the northeastern United States.
West Nile virus (WNV) has become firmly established in the Western Hemisphere since its discovery in the New York City area in 1999 (1,2). The virus has spread at an unprecedented rate throughout the continental United States and to neighboring countries, where it is maintained in an enzootic cycle that involves wild birds and ornithophilic mosquitoes (3). To date, 60 mosquito species have been found to be infected with WNV in North America; certain Culex spp. appear to be primary vectors, depending on region (4). In the northeastern United States, Culex pipiens, Cx. restuans, and Cx. salinarius have been implicated as the principal vectors because they are physiologically competent (5), frequently infected with the virus in nature, and closely associated with WNV transmission foci (6). However, the precise role that each of these species plays in enzootic transmission among birds or epidemic transmission to humans is not entirely clear.
Entomologic measures of risk may be estimated for different mosquito species by considering their abundance, biting behavior, prevalence of WNV infection, and vector competence. By synthesizing these parameters, Kilpatrick et al. (7) estimated that Cx. pipiens and Cx. restuans were responsible for up to 80% of human infections in New York, whereas Cx. salinarius accounted for only 4% of such infections. However, in Connecticut, the abundance of Cx. salinarius and prevalence of WNV infection in this species often approach those of Cx. pipiens (6). Observations in rural and urban sites in New York further indicate that Cx. pipiens and Cx. restuans are largely ornithophilic, whereas Cx. salinarius feeds more frequently on mammals (8), which supports the idea of a "bridge vector" role for this species. Nevertheless, collections from New Jersey indicate that mosquitoes of the Cx. pipiens complex may readily feed on mammals, including humans (9). Further blood meal analysis is required from mosquitoes collected in those habitats that support intense WNV transmission to more fully understand their respective roles as enzootic and epidemic vectors. Such information is vital to the success of any vector control program.
The current research initiative was undertaken to characterize the host-feeding patterns of Culex vectors and to evaluate their contribution to enzootic maintenance of WNV in wild bird populations and epidemic transmission to humans. Accordingly, blood-fed mosquitoes were collected from WNV transmission foci in Connecticut and analyzed for host source by sequencing polymerase chain reaction (PCR) amplification products of the vertebrate cytochrome b gene.
Mosquito Collection
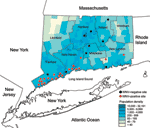
Mosquitoes were collected from 31 different sites in 6 counties in Connecticut during a 3-year period (June through October, 2002–2004) as part of a statewide surveillance program (6) and a focused trapping effort in Fairfield County (10). Most (71%) of the mosquito collection sites were located in densely populated residential locales along the urban/suburban corridor that extends from lower Fairfield and New Haven Counties, where high levels of WNV activity were recorded (Figure, Table 1). Trap sites included parks, greenways, golf courses, undeveloped wood lots, sewage treatment plants, dumping stations, and temporary wetlands associated with waterways. Three trap types were used: a CO2-baited CDC light trap (John W. Hock Co., Gainesville, FL, USA), a mosquito magnet experimental trap (American Biophysics Corp., East Greenwich, RI, USA), and a CDC gravid mosquito trap (11). Typically, traps were operated overnight and retrieved the following morning. Live, adult mosquitoes were transported to the laboratory, where they were promptly identified on chill tables with a stereomicroscope by using descriptive keys (12). All mosquitoes with fresh or visible blood remnants were transferred into individual 2-mL tubes labeled according to species, date of collection, and locale and stored at –80°C.
DNA Isolation from Blood-fed Mosquitoes
Mosquito abdomens were removed and reserved for blood-meal analysis with the aid of a dissecting microscope. Each mosquito was dissected individually on a new microscope slide by using flame-sterilized forceps to avoid cross-contamination. DNA was isolated from the abdominal contents of blood-fed mosquitoes individually by using DNA-zol BD, (Molecular Research Center, Cincinnati, OH, USA) according to the manufacturer's recommendation. Briefly, individual mosquito abdomens were homogenized with heat-sealed pipette tips in 1.5-mL tubes containing DNA-zol BD solution. The homogenates were incubated at room temperature for 5–10 min, mixed, and then centrifuged at 10,000 × g for 10 min. DNA was precipitated by adding isopropanol and 3–4 μL Poly Acryl Carrier (Molecular Research Center). The DNA pellet was then washed twice with 75% ethanol, air-dried briefly, reconstituted in TE buffer (10 mmol/L Tris-HCl, pH 8.0, 1 mmol/L EDTA) and stored at –20°C for further analysis.
Blood-meal Analysis
Isolated DNA from the mosquito blood meals served as DNA templates in subsequent PCRs as previously described (8,9). PCR primers were based either on a multiple alignment of cytochrome b sequences of avian and mammalian species obtained from GenBank or previously published primer sequences cited in Table 2. All DNA templates were initially screened with avian-a and mammalian-a primer pairs, and the sequences were analyzed (Table 2). In some cases, other primer pairs (avian b, mammalian b and c) were additionally used to resolve ambiguous sequences. A Taq PCR Core Kit (Qiagen, Germantown, MD, USA) was used for all PCRs according to the manufacturer's recommendation. A 50-μL reaction volume was prepared with 3 μmL template DNA, 4 μL each primer (0.1–0.5 μmol/L), 5 μL 10× Qiagen PCR Buffer (containing 15 mmol/L MgCl2), 1 μL dNTP mix (10 mmol/L each), 0.25 μL Taq DNA polymerase (1.25 U/reaction) and 32.75 μL water. All PCRs were performed with the GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA) at the ramp speed of 3°C–5°C/s. PCR-amplified products were purified by using QIAquick PCR Purification Kit (Qiagen) and sequenced directly in cycle-sequencing reactions at the Keck Sequencing Facility (Yale University, New Haven, CT, USA) by using the sequencer 3730xl DNA Analyzer (Applied Biosystems). Sequences were annotated by using ChromasPro version 1.22 (Technelysium Pty Ltd., Tewantin, Queensland, Australia) and identified by comparison to the GenBank DNA sequence database (13).
The performance of the molecular based assay was validated by isolating DNA from the blood of a number of known vertebrate species and subjecting it to PCR amplification and DNA sequencing. These species included American robin, American crow, black-capped chickadee, blue jay, button quail, common grackle, eastern tufted titmouse, gray catbird, house sparrow, mourning dove, northern cardinal, sharp-shinned hawk, wood thrush, domestic cat, domestic cow, domestic dog, horse, sheep, white-footed mouse, and white-tailed deer. Similar validation was also conducted with DNA isolated from blood-engorged, laboratory-reared Aedes aegypti that fed on guinea pig and button quail. Seasonal changes in the host feeding patterns of Cx. pipiens on selected host species were analyzed by χ2 analysis for trend by using GraphPad Instat version 3.0 for Windows (GraphPad Software, San Diego, CA, USA).
Blood-meal sources were successfully identified by DNA sequencing from 204 of 213 Cx. pipiens, 30 of 33 Cx. restuans, and 100 of 106 Cx. salinarius. Of 204 Cx. pipiens analyzed, 190 (93.1%) contained avian blood only, 5 (2.5%) mammalian, 1 (0.5%) amphibian, and 8 (3.9%) both avian and mammalian blood. Of 100 Cx. salinarius analyzed, 36 (36%) contained avian blood only, 53 (53%) mammalian, and 11 (11%) both avian and mammalian blood. All blood meals identified from Cx. restuans were avian-derived.
The composition of avian-derived blood meals is shown in Table 3. We identified 27 species of birds as hosts for Cx. pipiens; the most frequently identified species were American robin (40.4 % of avian and 37.7% of total), gray catbird (11.1% and 10.4%), and house sparrow (10.6% and 9.9%). Only 1 American crow–derived blood meal was identified for Cx. pipiens. Sixteen bird species were identified as hosts for Cx. restuans. American robin (36.7%) was the preferred host for Cx. restuans, as it was for Cx. pipiens, and no crow-derived blood meals were identified. We identified 13 species of birds as hosts for Cx. salinarius. The 2 most common avian species were black-capped chickadee (27.7% of avian and 11.7% of total) and American robin (25.5% and 10.8%). More crow-derived blood meals were identified (8.5% and 3.6%) in this mosquito species.
A seasonal shift from American robins to other avian species was noted with Cx. pipiens (Table 4). The χ2 test for linear trend showed that the proportion of American robin–derived blood meals decreased from June until October (p<0.0001). In June, 62.4% of all avian-derived blood meals were obtained from American robins, and this percentage declined to 26.7% in July and 38.9% in August. By September, 25.7% of the avian-derived blood meals were obtained from gray catbirds and 20.0% from mourning doves, while none was identified as being from American robins.
An analysis of the mammalian blood meal sources for Cx. pipiens and Cx. salinarius is shown in Table 5. We identified 10 host species for Cx. salinarius and 7 for Cx. pipiens. White-tailed deer (Odocoileus virginianus) was the most frequently identified host for Cx. salinarius (67.2% of mammalian and 38.7% of total). Human-derived blood meals were identified from 2 Cx. salinarius and 1 Cx. pipiens.
Our analysis on the blood-feeding behavior of Culex mosquitoes provides insight into their relative roles as enzootic and epidemic vectors of WNV in this region of the northeastern United States. We found that Cx. pipiens and Cx. restuans predominantly feed on avian hosts and focus their feeding activity on several key bird species that can support WNV transmission, in particular, American robins, gray catbirds, and house sparrows. By contrast, we found that Cx. salinarius feeds more opportunistically than Cx. pipiens and Cx. restuans and includes a relatively high proportion of mosquitoes with mixed blood meals from both avian and mammalian sources. This finding suggests that Cx. salinarius serves as a bridge vector by transferring WNV from viremic birds to mammalian hosts.
The preponderance of WNV isolations obtained from Cx. pipiens in surveillance activities conducted over the last 6 years (6,10,14,15) clearly incriminates this species as the predominant mosquito vector in this region. However, while enzootic transmission to birds is strongly supported by a number of host-preference studies on regional populations (8,9,16–18), no consensus has been reached on the role of Cx. pipiens in epidemic transmission of WNV to humans in the northeastern United States. Apperson et al. (9) recently identified mammalian-derived blood meals in 38% of blood-fed Cx. pipiens, 10.8% of which were human-derived (≈2.5% overall), collected from New Jersey. This finding led these researchers to conclude that Cx. pipiens was likely an epidemic vector in that region. This interpretation was viewed as consistent with the incidence of human cases in 3 densely populated urban areas of Connecticut in 2002, where most viral isolations (78%) were from Cx. pipiens (6). Kilpatrick et al. (7), integrating WNV testing data from New York from 2000 to 2003 with information on mosquito abundance, infection prevalence, vector competence, and biting behavior, further suggested that Cx. pipiens and Cx. restuans were responsible for up to 80% of human infections in that region. However, the validity of their conclusions was based on the identification of mammalian-derived blood in ≈19% of these 2 species and the assumption that humans were also included. Our analysis of blood meals from wild-caught female Cx. pipiens from established WNV foci in Connecticut is inconsistent with this supposition, as this species shows a strong tendency for avian blood and little inclination for mammalian hosts, including humans. We, therefore, conclude that while Cx. pipiens may occasionally feed on humans, it may not be the predominant vector of WNV to humans in our region of the northeastern United States. This finding is compatible with the lack of any mammalian-derived blood meals in blooded Cx. pipiens collected from suburban locales in nearby Westchester County, New York (9), but contrasts sharply with a recent study conducted in Delaware, where 69% of the blood meals taken by Cx. pipiens were from large mammals (19), which suggests a difference in host preference from more southern regions of its range.
Examination of the blood-fed mosquitoes in the present study showed an exclusively ornithophilic nature of Cx. restuans; all analyzed blood meals were from avian species. These findings were consistent with prior host preference studies (8,9,16,20) and strongly support the view that this predominant "early season" species is most likely involved in initiation and amplification of WNV transmission among wild birds and rarely, if ever, feeds on humans in this region. This finding differs from a recent blood-meal analysis by Gingrich and Williams (19), who found that a limited number (n = 9) of Cx. restuans from Delaware were highly mammalophilic (9:1 mammal-to-bird ratio). However, they concluded that this species was still primarily an enzootic vector since they never collected it in human landing collections.
Our findings regarding the blood-feeding patterns of Cx. salinarius reinforce those of previous studies (18,20–25) and indicate that this species feeds indiscriminately on both birds and mammals, including humans. By using separate PCR primer pairs for different vertebrate classes, we find that 11% of Cx. salinarius acquired blood meals from both avian and mammalian sources, versus ≈4% for Cx. pipiens. We cannot say whether all or most of these double-source blood meals represent multiple feeding episodes during the same gonotrophic cycle or the detection of residual DNA from a prior egg-laying cycle. However, mixed-source blood meals have been reported for a number of Culex species by using different methods for blood-meal identification (8,16). Regardless, our findings indicate that a relatively large fraction of the Cx. salinarius population readily feeds on both birds and mammals, which is a necessary condition for epidemic transmission to humans. The opportunistic feeding pattern of Cx. salinarius, in conjunction with its physiologic competence to transmit WNV (26), high infection rates in nature (10,14,15), and seasonal distribution that overlaps with human cases (6), all indicate that this species is a bridge vector of WNV to humans in the northeastern United States.
White-tailed deer were the single most important source of blood for Cx. salinarius in our study, which supports similar findings from New Jersey (9,18). The apparent affinity of Cx. salinarius for deer over other mammalian hosts is likely a function of deer's availability, as they are the most abundant large mammals in the region after humans. The role of deer in the ecology and transmission dynamics of WNV is unknown. Seroprevalence of WNV antibodies was 0%–6% among hunter-killed deer from New Jersey in 2001 (27), which suggests infrequent exposure to WNV relative to avian hosts, but frequency of exposure is still greater than that in humans (28). Widespread abundance of deer could be zooprophylactic by diverting feeding from avian amplifying hosts to deer. This possibility merits further study.
Several avian hosts are highly susceptible to WNV infection and can support viremia sufficient to infect culicine vectors. Reservoir competence values expressed as the duration and magnitude of infectious-level viremia were evaluated for 25 bird species and shown to be highest for passerine birds, including the blue jay, common grackle, house finch, American crow, house sparrow, and American robin (29). Field data further implicate a few species as reservoir hosts in northeastern United States on the basis of their exposure to WNV. When abundance and seroprevalence data were combined, house sparrows were estimated to be the most commonly infected bird species in New York City (30,31). These findings, combined with reservoir competence data, suggest that house sparrows are amplifying hosts in urban locales; however, other resident bird species, such as the northern cardinal, house finch, and gray catbird, were also frequently exposed to WNV (30). We show that in Connecticut, Cx. pipiens and Cx. restuans acquire blood meals predominately from American robins, implicating this species as a reservoir host for WNV. American robins are moderately competent and develop infectious-level viremia for a duration of ≈3 days (29). This species is most abundant in Connecticut from early spring to midsummer (32). Therefore, they may support more early- to mid-season (June–August) amplification of the virus. Our findings of a seasonal shift in Cx. pipiens from American robins to other avian species support this hypothesis.
We found that Cx. pipiens and Cx. restuans rarely fed upon American crows, despite their abundance (32) and high death rate from WNV infection throughout the region (33–35). Similar findings were reported from Cx. pipiens–complex mosquitoes collected from New York (8) and New Jersey (9). This finding suggests that American crows may also acquire WNV through other means in addition to mosquito transmission. American crows are susceptible to WNV infection by oral ingestion of the virus in aqueous solution and by eating infected bird carcasses (29). These birds are aggressive nest raiders and, therefore, could also acquire WNV infection by eating infected nestling birds. Transmission could also occur directly from bird to bird, as has been demonstrated in laboratory settings for this and other species (29,36,37). Our findings indicate that American crows may not be the primary amplifying hosts for infecting Culex mosquitoes with WNV in this region of the northeastern United States. Alternatively, we find that other common birds, including American robins, gray catbirds, and house sparrows, may play a greater role in supporting enzootic transmission.
Our PCR-based method took advantage of the conservation and diversity of mitochondrial sequences in identifying the source of vertebrate blood from mosquitoes. Mitochondrial DNA is a useful marker in phylogenetic studies and molecular systematics because of its maternal inheritance, haploid nature, and rapid rate of evolution (38). The cytochrome b gene, in particular, has successfully been used to identify taxonomic groups to the subspecies level and these sequences are publicly accessible from a wide array of different bird and mammal species in the GenBank database. By sequencing portions of the cytochrome b gene, we unambiguously identified the blood-meal source to the species level, which represents an improvement in sensitivity and specificity over earlier analyses.
Dr Molaei is a postdoctoral scientist at the Connecticut Agricultural Experiment Station. His current research interests include epidemiology of WNV, in particular mosquito-host interactions and host feeding patterns of mosquito vectors.
Acknowledgments
We thank Louis A. Magnarelli for providing vertebrate blood samples and John Shepard, Michael Thomas, Terrill Goodman, Michael Vasil, and our mosquito/arbovirus support group for collecting and identifying mosquitoes.
Funding for this research was provided by Laboratory Capacity for Infectious Disease Cooperative Agreement Number U50/CCU6806-01-1 from the Centers for Disease Control and Prevention, United States Department of Agriculture Specific Cooperative Agreement Number 58-6615-1-218, and Hatch Grant CONH00768.
References
- Anderson JF, Andreadis TG, Vossbrinck CR, Tirrell S, Wakem EM, French RA, Isolation of West Nile virus from mosquitoes, crows, and a Cooper's hawk in Connecticut. Science. 1999;286:2331–3. DOIPubMedGoogle Scholar
- Lanciotti R, Roehrig JT, Deubel V, Smith J, Parker M, Steele K, Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science. 1999;286:2333–7. DOIPubMedGoogle Scholar
- Komar N. West Nile virus: epidemiology and ecology in North America. Adv Virus Res. 2003;61:185–234. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. West Nile virus [homepage on the Internet]. 2005 Sep 14 [cited 2006 Jan 19]. Available from http://www.cdc.gov/ncidod/dvbid/westnile/mosquitoSpecies.htm
- Turell MJ, Dohm DJ, Sardelis MR, Oguinn ML, Andreadis TG, Blow JA. An update on the potential of North American mosquitoes (Diptera: Culicidae) to transmit West Nile virus. J Med Entomol. 2005;42:57–62. DOIPubMedGoogle Scholar
- Andreadis TG, Anderson JF, Vossbrinck CR, Main AJ. Epidemiology of West Nile virus in Connecticut: a five-year analysis of mosquito data 1999–2003. Vector Borne Zoonotic Dis. 2004;4:360–78. DOIPubMedGoogle Scholar
- Kilpatrick AM, Kramer LD, Campbell SR, Alleyne EO, Dobson AP, Daszak P. West Nile virus risk assessment and the bridge vector paradigm. Emerg Infect Dis. 2005;11:425–9.PubMedGoogle Scholar
- Apperson CS, Harrison BA, Unnasch TR, Hassan HK, Irby WS, Savage HM, Host-feeding habits of Culex and other mosquitoes (Diptera: Culicidae) in the borough of Queens in New York City, with characters and techniques for identification of Culex mosquitoes. J Med Entomol. 2002;39:777–85. DOIPubMedGoogle Scholar
- Apperson CS, Hassan HK, Harrison BA, Savage HM, Aspen SE, Farajollahi A, Host feeding patterns of established and potential mosquito vectors of West Nile virus in the eastern United States. Vector Borne Zoonotic Dis. 2004;4:71–82. DOIPubMedGoogle Scholar
- Anderson JF, Andreadis TG, Main AJ, Kline DL. Prevalence of West Nile virus in tree canopy-inhabiting Culex pipiens and associated mosquitoes. Am J Trop Med Hyg. 2004;71:112–9.PubMedGoogle Scholar
- Reiter P. A portable, battery-powered trap for collecting gravid Culex mosquitoes. Mosq News. 1983;43:496–8.
- Darsie RJ, Ward RA. Identification and geographic distribution of mosquitoes of North America, north of Mexico. Mosq Syst. 1981;1:1–313.
- The National Center for Biotechnology Information. GenBank. Available from http://www.ncbi.nlm.nih.gov/Genbank/index.html
- Andreadis TG, Anderson JF, Vossbrinck CR. Mosquito surveillance for West Nile virus in Connecticut, 2000: isolation from Culex pipiens, Cx. restuans, Cx. salinarius, and Culiseta melanura. Emerg Infect Dis. 2001;7:670–4. DOIPubMedGoogle Scholar
- Kulasekera VL, Kramer L, Nasci RS, Mostashari F, Cherry B, Trock SC, West Nile virus infection in mosquitoes, birds, horses, and humans, Staten Island, New York, 2000. Emerg Infect Dis. 2001;7:722–5. DOIPubMedGoogle Scholar
- Magnarelli LA. Host feeding patterns of Connecticut mosquitoes (Diptera: Culicidae). Am J Trop Med Hyg. 1977;26:547–52.PubMedGoogle Scholar
- Tempelis CH. Host-feeding patterns of mosquitoes, with a review of advances in analysis of blood meals by serology. J Med Entomol. 1975;11:635–53.PubMedGoogle Scholar
- Crans W. Continued host preference studies with New Jersey mosquitoes. Proceedings of the 51st annual meeting of the New Jersey Mosquito Exterminators Association; 1964. p. 50–8.
- Gingrich J, Williams GM. Host-feeding patterns of suspected West Nile virus mosquito vectors in Delaware, 2001–2002. J Am Mosq Control Assoc. 2005;21:194–200. DOIPubMedGoogle Scholar
- Irby WS, Apperson CS. Hosts of mosquitoes in the coastal plain of North Carolina. J Med Entomol. 1988;25:85–93.PubMedGoogle Scholar
- Edman JD. Host-feeding patterns of Florida mosquitoes. 3. Culex (Culex) and Culex (Neoculex). J Med Entomol. 1974;11:95–104.PubMedGoogle Scholar
- Cupp E, Stokes GM. Identification of bloodmeals from mosquitoes collected in light traps and dog-baited traps. Mosq News. 1973;33:39–41.
- Murphey F, Burbutis PP, Bray DF. Bionomics of Culex salinarius Coquillett. II. Host acceptance and feeding by adult females of Cx. salinarius and other mosquito species. Mosq News. 1967;27:366–74.
- Edman J, Downe AER. Host-blood sources and multiple habits of mosquitoes in Kansas. Mosq News. 1964;24:154–60.
- Cupp E, Stokes GM. Feeding patterns of Culex salinarius Coquillett in Jefferson Parish, Louisiana. Mosq News. 1976;36:332–5.
- Sardelis MR, Turell MJ, Dohm DJ, O'Guinn ML. Vector competence of selected North American Culex and Coquillettidia mosquitoes for West Nile virus. Emerg Infect Dis. 2001;7:1018–22. DOIPubMedGoogle Scholar
- Farajollahi A, Gates R, Crans W, Komar N. Serologic evidence of West Nile virus and St. Louis encephalitis virus infections in white-tailed deer (Odocoileus virginianus) from New Jersey, 2001. Vector Borne Zoonotic Dis. 2004;4:379–83. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Serosurveys for West Nile virus infection—New York and Connecticut counties, 2000. MMWR Morb Mortal Wkly Rep. 2001;50:37–9.PubMedGoogle Scholar
- Komar N, Langevin S, Hinten S, Nemeth N, Edwards E, Hettler D, Experimental infection of North American birds with the New York 1999 strain of West Nile virus. Emerg Infect Dis. 2003;9:311–22.PubMedGoogle Scholar
- Komar N, Burns J, Dean C, Panella NA, Dusza S, Cherry B. Serologic evidence for West Nile virus infection in birds in Staten Island, New York, after an outbreak in 2000. Vector Borne Zoonotic Dis. 2001;1:191–6. DOIPubMedGoogle Scholar
- Nasci RS, Komar N, Marfin AA, Ludwig GV, Kramer LD, Daniels TJ, Detection of West Nile virus-infected mosquitoes and seropositive juvenile birds in the vicinity of virus-positive dead birds. Am J Trop Med Hyg. 2002;67:492–6.PubMedGoogle Scholar
- Hanisek G. Connecticut birds by the season. The Connecticut Warbler. 2005;25:1–44.
- Hadler J, Nelson R, McCarthy T, Andreadis T, Lis MJ, French R, West Nile virus surveillance in Connecticut in 2000: an intense epizootic without high risk for severe human disease. Emerg Infect Dis. 2001;7:636–42. DOIPubMedGoogle Scholar
- Hochachka WM, Dhondt AA, McGowan KJ, Kramer LD. Impact of West Nile virus on American crows in the northeastern United States, and its relevance to existing monitoring program. EcoHealth. 2004;1:60–8. DOIGoogle Scholar
- Julian KG, Eidson M, Kipp AM, Weiss E, Petersen LR, Miller JR, Early season crow mortality as a sentinel for West Nile virus disease in humans, northeastern United States. Vector Borne Zoonotic Dis. 2002;2:145–55. DOIPubMedGoogle Scholar
- Komar N, Lanciotti R, Bowen R, Langevin S, Bunning M. Detection of West Nile virus in oral and cloacal swabs collected from bird carcasses. Emerg Infect Dis. 2002;8:741–2.PubMedGoogle Scholar
- McLean RG, Ubico SR, Docherty DE, Hansen WR, Sileo L, McNamara TS. West Nile virus transmission and ecology in birds. Ann N Y Acad Sci. 2001;951:54–7. DOIPubMedGoogle Scholar
- Moore W, DeFilipps VR. Taxonomic resolution based on cytochrome b DNA. In: Mindell DP, editor. Avian molecular evolution and systematics. San Diego: Academic Press; 1997.
- Cicero C, Johnson NK. Higher-level phylogeny of new world vireos (aves: vireonidae) based on sequences of multiple mitochondrial DNA genes. Mol Phylogenet Evol. 2001;20:27–40. DOIPubMedGoogle Scholar
- Sorenson M, Ast J, Dimcheff DE, Yuri T, Mindell DP. Primers for a PCR-based approach to mitochondrial genome sequencing in birds and other vertebrates. Mol Phylogenet Evol. 1999;12:105–14. DOIPubMedGoogle Scholar
Figure
Tables
Cite This ArticleTable of Contents – Volume 12, Number 3—March 2006
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Goudarz Molaei, The Connecticut Agricultural Experiment Station, 123 Huntington St, PO Box 1106, New Haven, CT 06504, USA; fax: 203-974-8502
Top