Volume 12, Number 5—May 2006
THEME ISSUE
Tuberculosis Special Section
Research
Beijing/W Genotype Mycobacterium tuberculosis and Drug Resistance
Abstract
Beijing/W genotype Mycobacterium tuberculosis is widespread, may be increasing, and may have a predilection for drug resistance. Individual-level data on >29,000 patients from 49 studies in 35 countries were combined to assess the Beijing genotype’s prevalence worldwide, trends over time and with age, and associations with drug resistance. We found 4 patterns for Beijing/W genotype tuberculosis (TB): 1) endemic, not associated with drug resistance (high level in most of East Asia, lower level in parts of the United States); 2) epidemic, associated with drug resistance (high level in Cuba, the former Soviet Union, Vietnam, and South Africa, lower level in parts of Western Europe); 3) epidemic but drug sensitive (Malawi, Argentina); and 4) very low level or absent (parts of Europe, Africa). This study confirms that Beijing/W genotype TB is an emerging pathogen in several areas and a predominant endemic strain in others; it is frequently associated with drug resistance.
The Mycobacterium tuberculosis genotype family known as "Beijing/W," "W-Beijing," or "Beijing" is widespread (1–3). Described in 1995 as the prevalent genotype in East Asia (4), >80% of strains from the Beijing area were of this type. The multidrug-resistant W strain is a member of the family. We use "Beijing" for the whole genotype family.
Researchers are concerned that the Beijing genotype may have a predilection for developing drug resistance (5) and may be spreading worldwide, perhaps as a result of increased virulence (6). A systematic review of the published literature in 2002 concluded that although Beijing genotype tuberculosis (TB) was widespread, associations with drug resistance varied, and little information on time trends was available (2).
The review highlighted the problems of relying on published literature: varying strain definitions; reporting bias; and limited information on selection criteria, population subgroups, age groups, or time trends. As part of the European Concerted Action on New Generation Genetic Markers and Techniques for the Epidemiology and Control of Tuberculosis, we have combined available datasets, using a common strain definition and individual-level data.
Studies for inclusion were identified from the systematic review and from contacting members of the European Concerted Action and authors of relevant articles published since the review. We aimed to include as many studies as possible in which the proportion of TB caused by the Beijing genotype could be ascertained in an unbiased way. Studies could represent all or random samples of patients in an area, hospital, or laboratory. Studies limited to outbreaks, drug-resistant isolates, or of <30 patients were excluded. A study description and individual patient data that included at least the year the case was diagnosed and the genotype were required.
Strain Classification
Three methods identify Beijing genotype strains: spoligotyping (7), IS6110 restriction fragment length polymorphism (RFLP) (8), and region A RFLP (9). The typical Beijing spoligotype shows hybridization to spacers 35–43. Beijing-like patterns with <9 spacers (but not solely spacers 37–38, which represents M. microti), were included (10).
By using IS6110 RFLP, fingerprints are compared to 19 patterns representative of the Beijing genotype (https://hypocrates.rivm.nl/bnwww/index.html). With standard techniques, allowing 1% position tolerance and classifying all matches >80% as Beijing, these patterns have 96%–100% sensitivity and 98%–100% specificity to detect Beijing strains, taking spoligotyping as the accepted standard (10). Sensitivity is increased by spoligotyping strains with RFLP patterns that match 75%–80% to the reference strains. The third technique uses a characteristic IS6110 insertion in region A. This method has 100% sensitivity and 98% specificity compared with spoligotyping (10).
Analysis
The proportion of Beijing genotype strains in each study was calculated overall and after excluding immigrants. The proportion of Beijing genotype in immigrants was examined by place of birth. Time trends were examined directly and by examining trends with age; an association with younger age groups would suggest that the proportion of TB attributable to the Beijing genotype was increasing. Associations with drug resistance were examined, after immigrants were excluded, with and without excluding patients with previous TB. For pooled analyses, heterogeneity in the associations between studies was examined, and the results presented are adjusted for study.
Data were received from 49 studies representing 29,259 TB patients in 35 countries, including 11 studies from the systematic review (2); other studies in the review had no individual patient data available, used nonstandard case definitions, or researchers declined to participate. Other studies were contributed by members or contacts of the Concerted Action or were identified from subsequently published studies. Details of all included studies are shown in Table A1.
The proportion of tuberculosis due to the Beijing genotype in the included studies is shown in Table A2.
Overall, 9.9% had the Beijing genotype. In Western Europe and the Czech Republic, the proportion was low: <6% of cases among nonimmigrants. In sub-Saharan Africa, the proportion was low except in Cape Town, South Africa. In Latin America, data were only available from Argentina and Brazil; both studies found <1% of TB cases were caused by Beijing genotype. In North America and the Caribbean, the proportion was higher (8%–14%). In the former Soviet Union the proportion was high: 45%–56% in Russia and 29% in Estonia. The proportion was low in India (1%), higher in Bangladesh (7%), and increased further east: >50% in many parts of Southeast and East Asia.
Analyses by region of birth showed similar patterns (Table 1). Beijing genotype strains were rare (0.5%) among immigrants from Eastern Europe other than the former Soviet Union; most came from the former Yugoslavia. The Beijing genotype was much less common among immigrants from the Indian subcontinent (3.4%) than among those from Southeast Asia (19%) or East Asia (58%). Beijing genotype strains were uncommon among immigrants from North Africa (3.0%), the Middle East (5.2%), and sub-Saharan Africa (2.2%, including 50 [2.1%] of 2,427 persons from Somalia). Among Middle Eastern immigrants, Beijing genotype was found in 6 (1%) of 620 persons from Turkey but in 8 (9.9%) of 81 from Afghanistan.
Time Trends
Time trends were analyzed among nonimmigrants within individual studies with >3 years of data (Table 2). (Studies from France, Iran, Thailand, Vietnam, and Spain are excluded because of small numbers in some years or absence of Beijing genotype strains).
All Western European sites except London showed a slight increase in Beijing strains over time, but this finding was only significant in the Netherlands. Combining data for Western Europe, the odds ratio (OR), adjusted for study, for having the Beijing genotype in the later period compared to the earlier period was 1.5 (95% confidence interval [CI] 1.2–1.9). This figure was unchanged after adjusting for age. The trend was similar after excluding the Netherlands (adjusted OR 1.7, 95% CI 0.96–3.1).
In St. Petersburg, Okayama, Buenos Aires, São Paulo, and San Francisco, no significant change occurred over time, but the studies only covered a few years. In Cape Town and Malawi, significant increases occurred over time and were unchanged after adjusting for age.
Trends with Age
Trends with age for studies with >3 cases of Beijing genotype TB among nonimmigrants are summarized in Table 3. Most Western European studies found the highest proportion of Beijing genotype TB in the youngest age groups. Overall, for Western Europe, compared to those age >50 years, the OR, adjusted for study, of having the Beijing genotype was 1.2 (0.87–1.6) for those 30–49 years of age, and 2.4 (95% CI 1.8–3.3) for those <30 years of age, ptrend<0.001. Excluding the Netherlands, the trend was stronger: adjusted OR 2.2 (95% CI 1.1–4.2) for those 30–49 years of age and 3.9 (95% CI 1.9–7.9) for those <30 years of age, ptrend<0.001.
In Russia and Estonia, Beijing genotype strains were more common in younger patients, and the trend was significant in St. Petersburg (p = 0.02). Overall, compared to those >50 years of age, the study-adjusted OR was 1.1 (95% CI 0.70–1.8) for those 30–49 years of age and 1.7 (95% CI 1.1–2.9) for those <30 years of age, ptrend = 0.02.
The African studies that found any Beijing strains noted a higher proportion in younger persons than in older persons. This difference was not significant in individual studies but was when studies were combined: adjusted OR, 1.9 (95% CI 1.1–3.4) for those 30–49 years of age and 2.1 (95% CI 1.2–3.7) for those <30 years of age, compared to those >50 years of age, ptrend = 0.03.
Among nonimmigrants in US studies, no significant trend occurred with age, either individually or overall. In Cuba, Beijing genotypes were more common in younger persons than in older persons in the larger study and overall (ptrend = 0.06). In Buenos Aires and São Paulo, all Beijing genotype–infected patients were <30 years of age (p = 0.002).
Most Asian studies showed no association with age, but trends were seen in Bangladesh, Vietnam, and Hong Kong. In Vietnam, Beijing genotype was more common in younger patients in all 4 studies: overall, compared to those >50 years of age, the study-adjusted OR was 1.5 (95% CI 1.0–2.2) for those 30–49 years of age and 2.7 (95% CI 1.7–4.2) for those <30 years of age, ptrend<0.001. In Hong Kong, Beijing genotypes were least common in patients <30 years of age.
Drug Resistance
Studies with drug resistance data for all or most patients and with >3 Beijing genotype TB patients among nonimmigrants are summarized in Table A3. In the Western European studies, with the exception of inner London, resistance was more common among Beijing genotype strains than among other strains. Beijing genotype was significantly associated with resistance in Denmark (rifampin and ethambutol), Finland (rifampin and streptomycin), and the Netherlands (streptomycin). Overall, the study-adjusted OR for the association of Beijing genotype and resistance among nonimmigrants in Western Europe was 1.8 (95% CI 1.2–2.7) for any drug, 1.7 (95% CI 0.95–2.9) for isoniazid, 4.0 (95% CI 1.4–11.9) for rifampin, 2.3 (95% CI 1.4–3.7) for streptomycin, 3.0 (95% CI 0.38–23.2) for ethambutol, and 4.2 (95% CI 1.2–14.7) for multidrug resistance (i.e., resistance to at least isoniazid and rifampin). Of the Western European studies, only those from Denmark, Hamburg, the Netherlands, and London had data on previous treatment. After patients who had previously received treatment were excluded, the associations in the Netherlands and Denmark persisted, and the adjusted combined ORs were similar to those overall but with wider CIs (e.g., 1.6, 95% CI 1.0–2.6 for any drug resistance).
In Russia and Estonia, Beijing genotype was strongly associated with resistance to all tested drugs. None of the patients in Estonia had been previously treated. In the Archangel Oblast, the association persisted after previously treated patients were excluded, but in St. Petersburg only the association with isoniazid resistance remained significant. In Cuba, Beijing genotype was associated with streptomycin resistance in both studies, and this association persisted after previously treated patients were excluded.
In Malawi and Zimbabwe, none of the Beijing genotype isolates was drug resistant. In Cape Town, 14 (35%) of the 40 Beijing isolates that were tested were drug resistant, but the resistance of most Beijing isolates and of the other isolates was unknown.
In the Asian studies, only those in Bangladesh, Vietnam, and Taiwan found more drug resistance in Beijing genotype strains. In Bangladesh, 99% of the patients had previously received treatment for TB. In Vietnam, the results were little changed by excluding the few previously treated patients. In Taiwan, previous treatment was unknown. Two studies found that Beijing genotypes were less commonly drug resistant. In China, Beijing genotypes were less likely to exhibit ethambutol resistance; no information was available on previous treatment. In Malaysia, among patients without previous treatment, 1 (2%) of 48 isolates from patients with the Beijing genotype and 33 (13%) of 252 isolates from patients with other genotypes were resistant to any drugs (p = 0.03).
Other Associations
In most studies, the proportion of nonimmigrants with the Beijing genotype was similar for men and women. In Japan, the proportion was higher among men, and in Malawi, it was higher among women. Only 23 studies had data on HIV status in nonimmigrants, and of these, 13 found no Beijing genotype, no HIV-positive patients, or information was lacking on HIV status of the patients with Beijing genotype. In the 10 remaining studies (inner London, Lyon, the Netherlands, Tuscany, San Francisco, Cuba [both studies], Buenos Aires, Malawi, and Ho Chi Minh City), no association was found between HIV status and Beijing genotype.
No significant association was found between strain type and site of tuberculosis (pulmonary or extrapulmonary) in any of the 20 studies in which this information was available and both types of tuberculosis were included. In Cuba, outside Havana, and in the Archangel Oblast patients with recurrent TB were more likely than patients in their first episode of disease to have the Beijing genotype, but these associations were lost after adjusting for drug resistance. No associations with previous TB were found in any of the other 17 studies for which information was available, but the numbers of recurrent cases were often small.
In this study, we have brought together published and unpublished data to document the spread of Beijing genotype tuberculosis worldwide. Little information was available from many countries including most of the Americas, Eastern Europe, North Africa, the Middle East, and Australasia. All eligible studies were requested, whether Beijing genotypes were found or not, and within the included studies, the proportion with Beijing genotype should be representative of those settings. The individual-level data allowed comparable analyses in all sites and pooled analysis within regions. This study complements the spoligotype database (3), which includes only studies that used spoligotyping and is more inclusive and less detailed epidemiologically. The database shows a similar global distribution of the Beijing genotype to that described here.
The proportion of TB attributable to the Beijing genotype is variable: high in Asia, apart from the Indian subcontinent, increasing further east; low in parts of Africa, Latin America, and Western Europe; intermediate in the United States and Cuba; low in Eastern Europe (other than the former Soviet Union); low in the Middle East (including <1% in a recent study from Tehran [11]). In Western Europe, Beijing genotype is more common among immigrant TB patients than among indigenous patients. The proportion of Beijing genotype TB among nonimmigrants may reflect the importance of immigrants to the total TB prevalence in these countries as well as the origin of these immigrants. Immigrants accounted for >50% of TB cases in London, the Netherlands, France, Denmark, Sweden, and Hamburg, compared to 25% of cases in Italy, 24% in Austria, 8% in Finland, and 4% in Spain.
Using information from time and age group trends, we found that an increasing proportion of TB is due to Beijing genotype strains in Western Europe, southern Africa, and the former Soviet Union. We found little evidence of increase in Asia, except in Vietnam and Bangladesh.
Strong associations with drug resistance have been found in the former Soviet Union, Cuba, and Vietnam. The combined data for Western Europe suggest an association there. No association was found in a large study in Malawi or in most of the Asian studies.
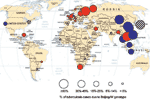
When the data on trends and drug resistance presented here and from other studies are combined, the results suggest that the distribution of Beijing genotype TB has several patterns (Figure). The Beijing genotype probably originated in the Beijing region of China (1,4); it was found in 90% of stored biopsy specimens in the 1950s, and this proportion has not changed over time (12). Beijing strains appear to have spread and become established as the predominant M. tuberculosis genotype in much of East and Southeast Asia, so little evidence of increase was found. In these areas, the Beijing genotype appears to be endemic and not associated with drug resistance (pattern 1).
In certain areas, including the former Soviet Union (13), Cuba, and Cape Town, epidemic spread was found, which was associated with drug resistance (pattern 2). Vietnam and Bangladesh follow this pattern, unlike most other Asian countries. Recent Indian studies suggest that India may also fit pattern 2 (14,15). In Taiwan, the association with drug resistance was not confirmed in a larger sample in 2003 (unpub. data), which suggests that it follows pattern 1. In parts of Western Europe, although the Beijing genotype remains uncommon, it appears to be increasing and is associated with drug resistance (pattern 2).
In the United States, the pattern is mixed. Nonimmigrant patients in San Francisco fit pattern 1: no association with drug resistance and no evidence of time trends. In this area, most Beijing isolates came from Asian immigrants, among whom no association was found between Beijing genotype and drug resistance. In the New Jersey study, no data on drug resistance were available, but a previous study in this area found that most Beijing isolates from nonimmigrants were pansusceptible (1,16). The age distribution does not suggest recent increase, which fits pattern 1. In contrast, the spread of the multidrug-resistant W strain in New York and beyond during the 1990s has been well documented (17–19). Other published studies from the United States confirm that the Beijing genotype is widespread but do not report drug resistance or trends (20–23).
In Malawi, an increase in the Beijing genotype over time was documented, but with drug sensitive strains (pattern 3). Argentina may fit this pattern, and spread of drug-sensitive Beijing genotype TB has been described in Gran Canaria (24). The final pattern (pattern 4) is of very low level or absent Beijing genotypes, as seen in parts of Africa and Europe.
The wide distribution of the Beijing genotype could be attributable to a founder effect or random drift, though these mechanisms would be unlikely to account for recent increases in multiple settings. The distribution could reflect particular stability of the genetic markers used to identify the genotype. High levels and epidemic spread may suggest that it transmits more easily or is more virulent than other strains. In vitro and animal studies have suggested increased multiplication or virulence for some Beijing strains (6,25) but not others (26). In Vietnam, the Beijing genotype was associated with treatment failure and relapse (27), but we found no such association. In Indonesia, patients with the Beijing genotype had a similar clinical picture to other TB patients for almost all parameters studied (28). In the Netherlands, the appearance on chest radiograph was similar for patients infected with Beijing genotype and for other TB patients (29). In Malawi, the Beijing genotype was not associated with death or transmissibility (30).
External factors may select for Beijing strains. In the former Soviet Union and the United States, spread has been associated with prisons and with high rates of drug resistance (13,17,31,32). In Mongolia, data were also available from prisoners. They had a higher proportion of Beijing genotype than did other patients, 46 (82%) of 56 compared to 97 (58%) of 168, p = 0.001, and a higher prevalence of drug resistance. Population movements (33), for example, from the former Soviet Union into Western Europe and through Afghanistan, may account for spread, recent increases, and the association with drug resistance (34).
Beijing genotypes may have a particular propensity to acquire drug resistance. Mutations in putative mutator genes have been found in Beijing genotypes, which suggests adaptability (5), but no increase in the rate of acquisition of resistance to rifampicin was found in in vitro studies (35). Once established, resistance could encourage spread if it delays effective treatment. Although the fitness of resistant strains is slightly reduced, this may be less marked for Beijing strains (36).
This study has confirmed that Beijing genotype M. tuberculosis is an emerging infection in many parts of the world and is a highly endemic pathogen in other areas. Its association with drug resistance, sometimes at high levels, in a number of settings, underlines its importance. The reasons for its apparent success are not well understood but may depend on human population movements as well as on any intrinsic factors.
Acknowledgments
We thank the many collaborators who contributed to this study. This study would not have been possible without the generous contribution of data from many studies.2
The European Concerted Action on New Generation Markers and Techniques for the Epidemiology and Control of Tuberculosis was funded by the European Union, grant QLK2-CT-2000-00630. J.R.G. was funded by the UK Department of Health, Public Health Career Scientist Award.
References
- Bifani PJ, Mathema B, Kurepina NE, Kreiswirth BN. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 2002;10:45–52. DOIPubMedGoogle Scholar
- Glynn JR, Whiteley J, Bifani PJ, Kremer K, van Soolingen D. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg Infect Dis. 2002;8:843–9.PubMedGoogle Scholar
- Filliol I, Driscoll JR, van Soolingen D, Kreiswith BN, Kremer K, Valetudie G, Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. J Clin Microbiol. 2003;41:1963–70. DOIPubMedGoogle Scholar
- Van Soolingen D, Qian L, de Haas PE, Douglas JT, Traore H, Portaels F, Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J Clin Microbiol. 1995;33:3234–8.PubMedGoogle Scholar
- Rad ME, Bifani P, Martin C, Kremer K, Samper S, Rauzier J, Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg Infect Dis. 2003;9:838–45.PubMedGoogle Scholar
- Lopez B, Aguilar D, Orozco H, Burger M, Espitia C, Ritacco V, A marked difference in pathogenesis and immune response induced by different Mycobacterium tuberculosis genotypes. Clin Exp Immunol. 2003;133:30–7. DOIPubMedGoogle Scholar
- Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–14.PubMedGoogle Scholar
- van Embden JDA, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–9.PubMedGoogle Scholar
- Kurepina NE, Sreevatsan S, Plikaytis BB, Bifani PJ, Connell ND, Donnelly RJ, Characterization of the phylogenetic distribution and chromosomal insertion sites of five IS6110 elements in Mycobacterium tuberculosis: non-random integration in the dnaA-dnaN region. Tuber Lung Dis. 1998;79:31–42. DOIPubMedGoogle Scholar
- Kremer K, Glynn JR, Lillebaek T, Niemann S, Kurepina NF, Kreiswirth BN, Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J Clin Microbiol. 2004;42:4040–9. DOIPubMedGoogle Scholar
- Farnia P, Mohammadi F, Masjedi MR, Varnerot A, Zarifi AZ, Tabatabee J, Evaluation of tuberculosis transmission in Tehran: using RFLP and spoligotyping methods. J Infect. 2004;49:94–101. DOIPubMedGoogle Scholar
- Qian L, van Embden JD, van Der Zanden AG, Weltevreden EF, Duanmu H, Douglas JT. Retrospective analysis of the Beijing family of Mycobacterium tuberculosis in preserved lung tissues. J Clin Microbiol. 1999;37:471–4.PubMedGoogle Scholar
- Drobniewski F, Balabanova Y, Ruddy M, Weldon L, Jeltkova K, Brown T, Rifampin- and multidrug-resistant tuberculosis in Russian civilians and prison inmates: dominance of the Beijing strain family. Emerg Infect Dis. 2002;8:1320–6.PubMedGoogle Scholar
- Singh UB, Suresh N, Bhanu NV, Arora J, Pant H, Sinha S, Predominant tuberculosis spoligotypes, Delhi, India. Emerg Infect Dis. 2004;10:1138–42.PubMedGoogle Scholar
- Almeida D, Rodrigues C, Ashavaid TF, Lalvani A, Udwadia ZF, Mehta A. High incidence of the Beijing genotype among multidrug-resistant isolates of Mycobacterium tuberculosis in a tertiary care center in Mumbai, India. Clin Infect Dis. 2005;40:881–6. DOIPubMedGoogle Scholar
- Bifani PJ, Mathema B, Liu Z, Moghazeh SL, Shopsin B, Tempalski B, Identification of a W variant outbreak of Mycobacterium tuberculosis via population-based molecular epidemiology. JAMA. 1999;282:2321–7. DOIPubMedGoogle Scholar
- Bifani PJ, Plikaytis BB, Kapur V, Stockbauer K, Pan X, Lutfey ML, Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA. 1996;275:452–7. DOIPubMedGoogle Scholar
- Moss AR, Alland D, Telzak E, Hewlett D Jr, Sharp V, Chiliade P, A city-wide outbreak of a multiple-drug-resistant strain of Mycobacterium tuberculosis in New York. Int J Tuberc Lung Dis. 1997;1:115–21.PubMedGoogle Scholar
- Agerton TB, Valway SE, Blinkhorn RJ, Shilkret KL, Reves R, Schluter WW, Spread of strain W, a highly drug-resistant strain of Mycobacterium tuberculosis, across the United States. Clin Infect Dis. 1999;29:85–92. DOIPubMedGoogle Scholar
- Barnes PF, Yang Z, Preston-Martin S, Pogoda MJ, Jones BE, Otaya M, Patterns of tuberculosis transmission in central Los Angeles. JAMA. 1997;278:1159–63. DOIPubMedGoogle Scholar
- Yang Z, Barnes PF, Chaves F, Eisenach KD, Weis SE, Bates JH, Diversity of DNA fingerprints of Mycobacterium tuberculosis isolates in the United States. J Clin Microbiol. 1998;36:1003–7.PubMedGoogle Scholar
- Soini H, Pan X, Amin A, Graviss EA, Siddiqui A, Musser JM. Characterization of Mycobacterium tuberculosis isolates from patients in Houston, Texas, by spoligotyping. J Clin Microbiol. 2000;38:669–76.PubMedGoogle Scholar
- Cowan LS, Crawford JT. Genotype analysis of Mycobacterium tuberculosis isolates from a sentinel surveillance population. Emerg Infect Dis. 2002;8:1294–302.PubMedGoogle Scholar
- Caminero JA, Pena MJ, Campos-Herrero MI, Rodriguez JC, Garcia I, Cabrera P. al. Epidemiological evidence of the spread of a Mycobacterium tuberculosis strain of the Beijing genotype on Gran Canaria Island. Am J Respir Crit Care Med. 2001;164:1165–70.PubMedGoogle Scholar
- Zhang M, Gong J, Yang Z, Samten B, Cave MD, Barnes PF. Enhanced capacity of a widespread strain of Mycobacterium tuberculosis to grow in human macrophages. J Infect Dis. 1999;179:1213–7. DOIPubMedGoogle Scholar
- Dormans J, Burger M, Aguilar D, Hernandex-Pando R, Kremer K, Roholl P, Correlation of virulence, lung pathology, bacterial load and delayed type hypersensitivity responses after infection with different Mycobacterium tuberculosis genotypes in a BALB/c mouse model. Clin Exp Immunol. 2004;137:460–8. DOIPubMedGoogle Scholar
- Lan NT, Lien HT. Tung le B, Borgdorff MW, Kremer K, van Soolingen D. Mycobacterium tuberculosis Beijing genotype and risk for treatment failure and relapse, Vietnam. Emerg Infect Dis. 2003;9:1633–5.PubMedGoogle Scholar
- van Crevel R, Nelwan RHH, de Lenne W, Veeraragu Y, van der Aznden AG, Armin Z, Mycobacterium tuberculosis Beijing genotype strains associated with febrile response to treatment. Emerg Infect Dis. 2001;7:1–4. DOIPubMedGoogle Scholar
- Borgdorff MW, van Deutekom H, de Haas PE, Kremer K, van Soolingen D. Mycobacterium tuberculosis, Beijing genotype strains not associated with radiological presentation of pulmonary tuberculosis. Tuberculosis (Edinb). 2004;84:337–40. DOIPubMedGoogle Scholar
- Glynn JR, Crampin AC, Traore H, Yates MD, Mwaungulu FD, Mgwira BM, Mycobacterium tuberculosis Beijing genotype, northern Malawi. Emerg Infect Dis. 2005;11:150–3.PubMedGoogle Scholar
- Pfyffer GE, Strassle A, van Gorkom T, Portaels F, Rigouts L, Mathieu C, Multidrug-resistant tuberculosis in prison inmates, Azerbaijan. Emerg Infect Dis. 2001;7:855–61. DOIPubMedGoogle Scholar
- Toungoussova OS, Mariandyshev A, Bjune G, Sandven P, Caugant DA. Molecular epidemiology and drug resistance of Mycobacterium tuberculosis isolates in the Archangel prison in Russia: predominance of the W-Beijing clone family. Clin Infect Dis. 2003;37:665–72. DOIPubMedGoogle Scholar
- van Helden PD, Warren RM, Victor TC, van der Spuy G, Richardson M, Hoal-van Helden E. Strain families of Mycobacterium tuberculosis. Trends Microbiol. 2002;10:167–8. DOIPubMedGoogle Scholar
- Kubica T, Rusch-Gerdes S, Niemann S. The Beijing genotype is emerging among multidrug-resistant Mycobacterium tuberculosis strains from Germany. Int J Tuberc Lung Dis. 2004;8:1107–13.PubMedGoogle Scholar
- Werngren J, Hoffner SE. Drug-susceptible Mycobacterium tuberculosis Beijing genotype does not develop mutation-conferred resistance to rifampin at an elevated rate. J Clin Microbiol. 2003;41:1520–4. DOIPubMedGoogle Scholar
- Toungoussova OS, Caugant DA, Sandven P, Mariandyshev AO, Bjune G. Impact of drug resistance on fitness of Mycobacterium tuberculosis strains of the W-Beijing genotype. FEMS Immunol Med Microbiol. 2004;42:281–90. DOIPubMedGoogle Scholar
- Lillebaek T, Andersen AB, Dirksen A, Glynn JR, Kremer K. Mycobacterium tuberculosis Beijing genotype. Emerg Infect Dis. 2003;9:1553–7.PubMedGoogle Scholar
- Puustinen K, Marjamaki M, Rastogi N, Sola C, Filliol I, Ruutu P, Characterization of Finnish Mycobacterium tuberculosis isolates by spoligotyping. J Clin Microbiol. 2003;41:1525–8. DOIPubMedGoogle Scholar
- Gutierrez MC, Vincent V, Aubert D, Bizet J, Gaillot O, Lebrun L, Molecular fingerprinting of Mycobacterium tuberculosis and risk factors for tuberculosis transmission in Paris, France, and surrounding area. J Clin Microbiol. 1998;36:486–92.PubMedGoogle Scholar
- Diel R, Schneider S, Meywald-Walter K, Ruf CM, Rusch-Gerdes S, Niemann S. Epidemiology of tuberculosis in Hamburg, Germany: long-term population-based analysis applying classical and molecular epidemiological techniques. J Clin Microbiol. 2002;40:532–9. DOIPubMedGoogle Scholar
- Bonora S, Gutierrez MC, Di Perri G, Brunello F, Allegranzi B, Ligozzi M, Comparative evaluation of ligation-mediated PCR and spoligotyping as screening methods for genotyping of Mycobacterium tuberculosis strains. J Clin Microbiol. 1999;37:3118–23.PubMedGoogle Scholar
- Lari N, Rindi L, Bonanni D, Tortoli E, Garzelli C. Beijing/W Mycobacterium tuberculosis in Italy. Emerg Infect Dis. 2004;10:958–9.PubMedGoogle Scholar
- Borgdorff MW, de Haas P, Kremer K, van Soolingen D. Mycobacterium tuberculosis Beijing genotype, the Netherlands. Emerg Infect Dis. 2003;9:1310–3.PubMedGoogle Scholar
- Ruiz Garcia M, Rodriguez JC, Navarro JF, Samper S, Martin C, Royo G. Molecular epidemiology of tuberculosis in Elche, Spain: a 7-year study. J Med Microbiol. 2002;51:273–7.PubMedGoogle Scholar
- Samper S, Iglesias MJ, Rabanaque MJ, Lezcano MA, Vitoria LA, Rubio MC, The molecular epidemiology of tuberculosis in Zaragoza, Spain: a retrospective epidemiological study in 1993. Int J Tuberc Lung Dis. 1998;2:281–7.PubMedGoogle Scholar
- Brudey K, Gordon M, Mostrom P, Svensson L, Jonsson B, Sola C, Molecular epidemiology of Mycobacterium tuberculosis in western Sweden. J Clin Microbiol. 2004;42:3046–51. DOIPubMedGoogle Scholar
- Hayward AC, Goss S, Drobniewski F, Saunders N, Shaw RJ, Goyal M, The molecular epidemiology of tuberculosis in inner London. Epidemiol Infect. 2002;128:175–84. DOIPubMedGoogle Scholar
- Maguire H, Dale JW, McHugh TD, Butcher PD, Gillespie SH, Costetsos A, Molecular epidemiology of tuberculosis in London 1995–7 showing low rate of active transmission. Thorax. 2002;57:617–22. DOIPubMedGoogle Scholar
- Kurepina NE, Kreiswirth BN, Shaskina E, Driscoll JR, Polanecky V, Kozakova , Molecular epidemiology of tuberculosis in Prague and South Moravia, Czech Republic: genetic analysis of Mycobacterium tuberculosis isolates by IS6110-RFLP fingerprinting and spoligotyping. Cent Eur J Public Health. 2004;12:141–50.PubMedGoogle Scholar
- Kruuner A, Hoffner SE, Sillastu H, Danilovits M, Levina K, Svenson SB, Spread of drug-resistant pulmonary tuberculosis in Estonia. J Clin Microbiol. 2001;39:3339–45. DOIPubMedGoogle Scholar
- Narvskaya O, Mokrousov I, Limeschenko E, Otten T, Steklova L, Graschenkova O, Molecular characterisation of Mycobacterium tuberculosis strains from the northwest region of Russia. EpiNorth. 2000 [cited 2006 Mar 27]. Available from http://www.epinorth.org/eway/default0.asp?pid=230&oid=0&e=0&trg=MainArea_5260&MainArea_5260=5263:0:15,2930:1:0:0:5260;::0:0:0
- Mokrousov I, Otten T, Vyshnevskiy B, Narvskaya O. Detection of embB306 mutations in ethambutol-susceptible clinical isolates of Mycobacterium tuberculosis from Northwestern Russia: implications for genotypic resistance testing. J Clin Microbiol. 2002;40:3810–3. DOIPubMedGoogle Scholar
- Toungoussova OS, Sandven P, Mariandyshev AO, Nizovtseva NI, Bjune G, Caugant DA. Spread of drug-resistant Mycobacterium tuberculosis strains of the Beijing genotype in the Archangel Oblast, Russia. J Clin Microbiol. 2002;40:1930–7. DOIPubMedGoogle Scholar
- Doroudchi M, Kremer K, Basiri EA, Kadivar MR, Van Soolingen D, Ghaderi AA. IS6110-RFLP and spoligotyping of Mycobacterium tuberculosis isolates in Iran. Scand J Infect Dis. 2000;32:663–8. DOIPubMedGoogle Scholar
- Niobe-Eyangoh SN, Kuaban C, Sorlin P, Cunin P, Thonnon J, Sola C, Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J Clin Microbiol. 2003;41:2547–53. DOIPubMedGoogle Scholar
- Kuaban C, Bercion R, Noeske J, Cunin P, Nkamsse P, Ngo Niobe S. Anti-tuberculosis drug resistance in the West Province of Cameroon. Int J Tuberc Lung Dis. 2000;4:356–60.PubMedGoogle Scholar
- Bruchfeld J, Aderaye G, Palme IB, Bjorvatn B, Ghebremichael S, Hoffner S, Molecular epidemiology and drug resistance of Mycobacterium tuberculosis isolates from Ethiopian pulmonary tuberculosis patients with and without human immunodeficiency virus infection. J Clin Microbiol. 2002;40:1636–43. DOIPubMedGoogle Scholar
- Kallenius G, Koivula T, Ghebremichael S, Hoffner SE, Norberg R, Svensson E, Evolution and clonal traits of Mycobacterium tuberculosis complex in Guinea-Bissau. J Clin Microbiol. 1999;37:3872–8.PubMedGoogle Scholar
- Glynn JR, Crampin AC, Traore H, Yates MD, Mwaungulu F, Ngwira B, Mycobacterium tuberculosis Beijing genotype, northern Malawi. Emerg Infect Dis. 2005;11:150–3.PubMedGoogle Scholar
- van Rie A, Warren RM, Beyers N, Gie RP, Classen CN, Richardson M, Transmission of a multidrug-resistant Mycobacterium tuberculosis strain resembling "strain W" among noninstitutionalized, human immunodeficiency virus-seronegative patients. J Infect Dis. 1999;180:1608–15. DOIPubMedGoogle Scholar
- Sharaf-Eldin GS, Saeed NS, Hamid ME, Jordaan AM, Van der Spuy GD, Warren RM, Molecular analysis of clinical isolates of Mycobacterium tuberculosis collected from patients with persistent disease in the Khartoum region of Sudan. J Infect. 2002;44:244–51. DOIPubMedGoogle Scholar
- Easterbrook PJ, Gibson A, Murad S, Lamprecht D, Ives N, Ferguson A, High rates of clustering of tuberculosis strains in Harare, Zimbabwe: a molecular epidemiological study. J Clin Microbiol. 2004;42:4536–44. DOIPubMedGoogle Scholar
- Bifani PJ, Mathema B, Liu Z, Moghazeh S, Shopsin B, Tempalski B, Identification of a W variant outbreak of Mycobacterium tuberculosis via population-based molecular epidemiology. JAMA. 1999;282:2321–7. DOIPubMedGoogle Scholar
- Jasmer RM, Hahn JA, Small PM, Daley CL, Behr MA, Moss AR, A molecular epidemiologic analysis of tuberculosis trends in San Francisco, 1991-1997. Ann Intern Med. 1999;130:971–8.PubMedGoogle Scholar
- Diaz R, Kremer K, de Haas PE, Gomez RI, Marrero A, Valdivia JA, Molecular epidemiology of tuberculosis in Cuba outside of Havana, July 1994-June 1995: utility of spoligotyping versus IS6110 restriction fragment length polymorphism. Int J Tuberc Lung Dis. 1998;2:743–50.PubMedGoogle Scholar
- Diaz R, Gomez R, Restrepo E, Rumbaut R, Sevy-Court J, Valdivia JA, Transmission of tuberculosis in Havana, Cuba: a molecular epidemiological study by IS6110 restriction fragment length polymorphism typing. Mem Inst Oswaldo Cruz. 2001;96:437–43. DOIPubMedGoogle Scholar
- Bhanu NV, van Soolingen D, van Embden JD, Dar L, Pandey RM, Seth P. Predominace of a novel Mycobacterium tuberculosis genotype in the Delhi region of India. Tuberculosis (Edinb). 2002;82:105–12. DOIPubMedGoogle Scholar
- Shamputa IC, Rigouts L, Eyongeta LA, El Aila NA, Van Deun A, Salim AH, Genotypic and phenotypic heterogeneity among single M tuberculosis isolates from pulmonary tuberculosis patients. J Clin Microbiol. 2004;42:5528–36. DOIPubMedGoogle Scholar
- van Crevel R, Nelwan RHH, de Lenne W, Veeraragu Y, van der Zanden AG, Amin Z, Mycobacterium tuberculosis Beijing genotype strains associated with febrile response to treatment. Emerg Infect Dis. 2001;7:880–3. DOIPubMedGoogle Scholar
- Dale JW, Nor RM, Ramayah S, Tang TH, Zainuddin ZF. Molecular epidemiology of tuberculosis in Malaysia. J Clin Microbiol. 1999;37:1265–8.PubMedGoogle Scholar
- Prodinger WM, Bunyaratvej P, Prachaktam R, Pavlic M. Mycobacterium tuberculosis isolates of Beijing genotype in Thailand. Emerg Infect Dis. 2001;7:483–4.PubMedGoogle Scholar
- Anh DD, Borgdorff M, Van LN, Lan NTN, van Gorkom T, Kremer K, Mycobacterium tuberculosis Beijing genotype emerging in Vietnam. Emerg Infect Dis. 2000;6:302–5. DOIPubMedGoogle Scholar
- Chan MY, Borgdorff M, Yip CW, de Haas PE, Wong WS, Kam KM, Seventy percent of the Mycobacterium tuberculosis isolates in Hong Kong represent the Beijing genotype. Epidemiol Infect. 2001;127:169–71. DOIPubMedGoogle Scholar
- Ohata R, Tada A. IS6110 restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolated in Okayama Prefecture [in Japanese]. Kekkaku. 2002;77:629–37.PubMedGoogle Scholar
- Naranbat N, Kremer K, Nymadawa P, Van Soolingen D. Abstract 101: Distribution the Beijing genotype of Mycobacterium strains isolated in Mongolia. In: 22nd International Union Against Tuberculosis and Lung Disease Eastern Region Conference, Kathmandu, Nepal, 2003 Sept 23–25. Abstract 101.
- Jou R, Chiang C-Y, Huang W-L. Distribution of the Bejing family genotypes of Mycobacterium tuberculosis in Taiwan. J Clin Microbiol. 2005;43:95–100. DOIPubMedGoogle Scholar
Figure
Tables
Cite This Article1Analysis and writing committee: Judith R. Glynn, London School of Hygiene and Tropical Medicine, London, UK; Kristin Kremer, RIVM, Bilthoven, the Netherlands; Martien W. Borgdorff, Royal Netherlands Tuberculosis Association (KNCV) Tuberculosis Foundation, The Hague, the Netherlands; Mar Pujades Rodriguez, London School of Hygiene and Tropical Medicine, London, UK; and Dick van Soolingen, RIVM, Bilthoven, the Netherlands.
2The key contacts who contributed the data are listed as follows: Austria: Wolfgang Prodinger (Medizinische Universität Innsbruck); Denmark: Troels Lillebaek (Statens Serum Institut, Copenhagen); Finland: Hanna Soini, Petri Ruutu, (National Public Health Institute, Helsinki); France: Cristina Gutierrez, Veronique Vincent (Institut Pasteur, Paris); Beate Heym, Veronique Friocourt (Hôpital Ambroise Paré, Boulogne-Billancourt); Isabelle Fredenucci, Jean-Pierre Flandrois (Centre Hospitalier Lyon-Sud, Lyon); Germany: Stefan Niemann (National Reference Centre for Mycobacteria, Forschungszentrum Borstel, Hamburg), Roland Diel (School of Public Health, University of Düsseldorf); Italy: Stefano Bonora (Università di Verona); Leonardo A Sechi, Stephania Zanetti (Università di Sassari); Carlo Garzelli (Università di Pisa); the Netherlands: Martien Borgdorff (KNCV Tuberculosis Foundation) Petra de Haas, Kristin Kremer, Dick van Soolingen (RIVM); Spain: Montserrat Ruiz, Juan Carlos Rodríguez, Gloria Royo (Universidad Miguel Hernández, Elche); Ana Pérez Meixeira, Jenaro Astray (Public Health Institute Getafe, Madrid), Juana Cacho, Amador Ramos (Hospital Universitario de Getafe); Maria Jose Iglesias (University of Zaragoza), Sofia Samper (Hospital Universitario Miguel Servet, Zaragoza); United Kingdom: Andrew Hayward, John Watson, Francis Drobniewski (Health Protection Agency, London); Jeremy Dale (University of Surrey) on behalf of the Steering Committee, Molecular Epidemiology of Tuberculosis in London; Sweden: Malin Ridell, Liselott Svensson (Institute of Medical Microbiology and Immunology, Göteborg University); Czech Republic: Milan Kubin (Institute of Hygiene of the City of Prague); Estonia: Annika Krüüner (Tartu University, Estonia, and Karolinska Institute, Stockholm, Sweden); Russia: Olga Toungoussova (University of Oslo, Norway), Dominique Caugant (Norwegian Institute of Public Health, Oslo, Norway), Andrey Mariandyshev (Northern State Medical University, Archangel): Olga Narvaskaya, Igor Mokrousov (St. Petersburg Pasteur Institute), Tatjana Otten, Boris Vyshnevskiy (Research Institute of Phthisiopulmonology, St. Petersburg); Iran: Mehrnoosh Doroudchi (Shiraz University of Medical Sciences); Cameroon: Sara Ngo Niobe-Eyangoh (Centre Pasteur du Cameroun, Yaoundé); Ethiopia: Judith Bruchfeld (Swedish Institute for Infectious Disease Control, Solna); Guinea Bissau: Tuija Koivula, Gunilla Kallenius (Swedish Institute for Infectious Disease Control, Solna); Malawi: Amelia Crampin, Judith Glynn (London School of Hygiene and Tropical Medicine, UK) on behalf of The Karonga Prevention Study (Chilumba, Malawi); South Africa: Madalene Richardson, Paul van Helden, Rob Warren, Nulda Beyers (Stellenbosch University, Cape Town); Sudan: Ghada Sharaf-Eldin (National Health Laboratory, Khartoum); Zimbabwe: Philippa Easterbrook, Shahed Murad, Francis Drobniewski (King’s College London, UK); Cuba: Raul Diaz (Instituto Pedro Kourí, Havana); United States: Barry Kreiswirth (International Center for Public Health, Newark, NJ); Midori Kato-Maeda, Elizabeth Fair, Sebastien Gagneux, Peter Small (Stanford University, Stanford, CA); Argentina: Nora Morcillo (Reference Laboratory of Buenos Aires Tuberculosis Control Program) Angel Cataldi (National Institute of Agricultural Technology); Brazil: Lucilaine Ferrazoli (Instituto Adolfo Lutz, Sao Paulo); India: Kristin Kremer (RIVM), P. Seth (All India Institute of Medical Sciences, New Delhi); Bangladesh: Leen Rigouts, Isdore Chola Shamputa (Institute of Tropical Medicine, Antwerp, Belgium); Indonesia: Reinout van Crevel (University Medical Center Nijmegen, the Netherlands); Malaysia: Jeremy Dale (University of Surrey, Guildford, UK); Thailand: Wolfgang Prodinger (Medizinische Universität Innsbruck, Austria), Porntip Bunyaratevej (Mahidol University, Bangkok); China: James Douglas (University of Hawaii); Li Weimin (Beijing Tuberculosis and Chest Tumor Institution), Kristin Kremer (RIVM); K.M. Kam (Tuberculosis Reference Laboratory, Hong Kong); Japan: Ritsuko Ohata (Okayama Prefectural Institute for Environmental Science and Public Health); Mongolia: N. Naranbat (National Center for Communicable Diseases, Ulaanbaatar); Vietnam: Dang Duc Anh (National Institute of Hygiene and Epidemiology, Hanoi); Mai Huyen, Nguyen Thi Ngoc Lan (Ho Chi Minh City); Taiwan: Ruwen Jou (Center for Disease Control, Taipei).
Table of Contents – Volume 12, Number 5—May 2006
| EID Search Options |
|---|
|
|
|
|
|
|