Volume 13, Number 6—June 2007
Research
Isolation and Characterization of Novel Human Parechovirus from Clinical Samples
Figure
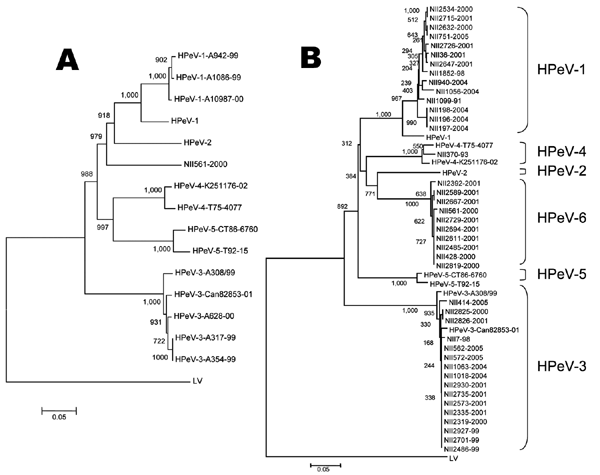
Figure. Phylogenetic tree analysis of the NII561-2000 virus and the NII561-2000–related viruses. A) The phylogenetic tree of P1 amino acid sequences was constructed as described in Materials and Methods. Bar shows a genetic distance of 0.05. The following amino acid sequences were obtained from GenBank: L02971 for HPeV-1 (Harris strain), AJ005695 for HPeV-2 (Williamson strain), AB084913 for HPeV-3 (A308/99), AJ889918 for HPeV-3 (Can82853-01), DQ315670 for HPeV-4 (K251176-02), AM235750 for HPeV-4 (T75-4077), AF055846 for HPeV-5 (CT86-6760), AM235749 for HPeV-5 (T92-15), and AF020541 for Ljungan virus (LV). B) Phylogenetic relationships among the NII561-2000 virus, the NII561-2000–related viruses, and other prototype HPeVs based on comparisons of amino acid similarities of capsid protein (the first 200 aa of VP0 sequences). The tree was constructed by the neighbor-joining method, using ClustalX. Bootstrap values are expressed in percentages after sampling 1,000×. Bar shows a genetic distance of 0.05.