Volume 13, Number 6—June 2007
Dispatch
Reemergence of Oropouche Fever, Northern Brazil
Cite This Article
Citation for Media
Abstract
Oropouche fever has reemerged in Parauapebas and Porto de Moz municipalities, Pará State, Brazil. Serologic analysis (immunoglobulin M–ELISA) and virus isolation confirmed Oropouche virus (OROV) in both municipalities. Nucleotide sequencing of 2 OROV isolates from each location indicated genotypes I (Parauapebas) and II (Porto de Moz) in Brazil.
Oropouche virus (OROV), the cause of Oropouche fever, belongs to the family Bunyaviridae, genus Orthobunyavirus, Simbu serogroup (1), and is transmitted between humans in urban areas by the biting midge Culicoides paraensis (2,3). This virus was first isolated from febrile forest workers in Trinidad in 1955. The first isolation in Brazil was in 1960 from the blood of a sloth (Bradypus tridactylus) (4). The epidemic potential of OROV was recognized during an outbreak in Belém, Pará State, Brazil, in 1961, where ≈11,000 persons were infected (4). Over the past 45 years, many outbreaks of Oropouche fever, ≈500,000 cases, have been described in the Americas. OROV has been isolated in Trinidad, Panama, Peru, and Brazil, and in the past 40 years Oropouche fever has emerged as a public health problem in tropical areas of Central and South America (3).
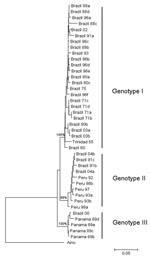
Members of the genus Orthobunyavirus have a tripartite, single-stranded, negative-sense RNA genome of small (S), medium (M), and large (L) RNAs that encode nucleocapsid, glycoproteins, and RNA polymerase, respectively. Phylogenetic analysis of nucleocapsid genes of different OROV strains identified 3 distinct genotypes (I, II, and III) currently circulating in Central and South America; genotypes I and II have been detected in the Brazilian Amazon (5). Recently, an OROV isolate from a marmoset (Callithrix sp.) was characterized as a member of genotype III (6).
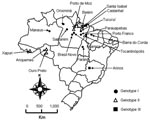
Two outbreaks of Oropuche fever occurred during 2003 and 2004. The first occurred in April–May 2003 in 2 communities (Vila Sansão, 140 inhabitants, and Vila Paulo Fontelles, 835 inhabitants).in the municipality of Parauapebas (6°4′S, 49°54′W). The second outbreak occurred in July–August 2004 in 1 community (Vila Tapara, 2,000 inhabitants) in the municipality of Porto de Moz (1°45′S, 52°14′W) (Figure 1).
A total of 125 and 109 serum samples were collected from residents of Parauapebas and Porto de Moz, which represented 12.8% and 5.45% of all inhabitants, respectively. Criteria for sampling were a history of acute fever several weeks before or during the survey or clinical symptoms similar to those of Oropouche fever. All serum samples were analyzed by hemagglutination inhibition (HI) test (7) and immunoglobulin M–ELISA (8) for specific HI and IgM antibodies to OROV. HI titers >20 and ELISA results greater than the cut-off value (optical density >0.200) were considered positive (8).Virus isolation was conducted by intracranial injection of newborn mice with a 1:10 (v/v) suspension of serum samples in phosphate-buffered saline, pH 7.4, as described elsewhere (9). Fifty-four and 11 serum samples from Parauapebas and Porto de Moz, respectively, were used for virus isolation. Identification of isolates was performed by complement fixation test as reported (9). Two OROV strains were isolated from patients in Parauapebas, and 2 strains were isolated from patients in Porto de Moz.
To genetically characterize the viruses, 2 isolates were selected from Parauapebas (Brazil 2003a and Brazil 2003b) and 2 from Porto de Moz (Brazil 2004a and Brazil 2004b). Viral RNA was extracted from Vero cells infected with human samples, and S RNA was amplified by using a 1-step reverse transcription–PCR assay as described (5,6). Phylogenetic trees were constructed for nucleocapsid gene nucleotide sequences by comparison with other OROV nucleocapsid gene sequences in GenBank (Table 1); neighbor-joining analysis (10) implemented in Mega version 2.1 (11) was used. Bootstrap analyses were performed on 1,000 replicates to generate confidence for groupings (12).
Of 125 serum samples from patients in Parauapebas, HI results were positive for 16 (12.7%) from Vila Sansão, 6 (4.8%) from Paulo Fontelles, and 4 (3.2%) from other localities. IgM was detected in 16 (12.7%), 8 (4.8%), and 6 (4.8%) serum samples from these 3 areas, respectively. Of 117 serum samples from patients in Porto de Moz, 56 (46.7%) had HI antibodies and 61 (52.1%) had IgM to OROV.
A total of 71.9% of female patients in Parauapebas and 59% in Porto de Moz had symptoms suggestive of Oropouche fever. Although all age groups were affected, persons 5–14 years of age had the highest frequency of symptoms (30.4%) and those <1–4 years of age had the lowest frequency (4.8%) (Table 2). Symptoms most frequently reported were fever (100%), headache (79.3%), joint pain (68.7%), and muscle pain (30%). Seventy percent of patients reported ≥1 episode of recurrence of fever, characterized by fever, headache, and other symptoms ≈2–3 weeks after onset of initial symptoms (2,3).
Full-length S RNA of the 4 OROV strains contained 754 nt and encoded 2 overlapping open reading frames, the nucleocapsid (693 nt and 231 aa) and nonstructural protein (273 nt and 91 aa). Two small noncoding regions were also found at the 3′ and 5′ ends of these reading frames, spanning nt positions 1–44 and 741–754, respectively. Phylogenetic analysis of Brazil 2003 and 2004 isolates grouped strains from Parauapebas (Brazil 2003a and Brazil 2003b) into OROV genotype I and strains from Porto de Moz (Brazil 2004a and Brazil 2004b) into OROV genotype II (Figure 2).
Oropouche fever is the second most common arboviral disease (after dengue fever) in the Brazilian Amazon region. From 1960 to 1980, Oropouche fever outbreaks were detected only in Pará State, mainly in Belém and neighboring areas, where thousands of people were infected (2,3). OROV was then detected in other Amazonian states including Amazonas, Amapá, Acre, Rondônia, and Tocantins; and non-Amazonian states, including Maranhão in northeastern Brazil and Tocantins in central Brazil (3,8). Recently, OROV isolated from Callithrix sp.in Arinos, Minas Gerais State, southeastern Brazil was characterized as genotype III, which indicated the presence of this genotype in Brazil (6). OROV from this species has been identified only in Panama (5). From 1980 to 2005, sporadic cases or self-limited outbreaks of Oropouche fever were reported in areas of the Brazilian Amazon, which suggested silent endemic circulation of the virus (13). In 2003 and 2004, several cases of Oropouche fever were detected in Parauaebas and Porto de Moz in Pará State. Parauaebas is located in the Carajás mineral province and Porto de Moz is located in the Altamira region.
Genetic characterization of strains indicated the presence of genotype II in the eastern Amazon region. This genotype had been associated with cases of Oropouche fever in restricted western Amazonian areas (Rondônia State), as well as in Peru (5). This finding suggests movement of OROV genotype II across the Amazon region from western to eastern areas or emergence of this genotype after silent circulation for several years. Genotype I (Brazil 2003a and Brazil 2003b) found in Parauapebas was closely related to Trinidadian and Brazilian isolates obtained from 1955 through 1960 (Trinidad 55 and Brazil 60) (5). Genotype II strains isolated in Porto de Moz were genetically related to strains isolated in Peru during the 1990s (Peru 92, 93, 97, 98a, 98b) and Rondônia State in 1991 (Brazil 91a, 91b), as reported by Saeed et al. (5). These data indicate that Parauapebas and Porto de Moz OROV isolates are genetically distinct and have different ancestor viruses (Figure 2). Recognition of different OROV genotypes in the Brazilian Amazon, as well as new genetic information, is useful for understanding the epidemiology and genetic diversity of this emergent human pathogen.
Dr Azevedo is a physician in the Department of Arbovirology and Hemorrhagic Fever at the Instituto Evandro Chagas, Ministry of Health, Belém, Pará State, Brazil. Her research interests include clinical, epidemiologic, and experimental studies of arboviruses, particularly those responsible for illness in humans.
Acknowledgments
We thank Alan Barrett for his invaluable comments and suggestions; Vanúsia Dias Duarte, Márcia Solange Ferro de Melo Silva, Núbia Maria de Lima Costa, and Solange Medeiros da Silva for their assistance; and Basílio Silva Buna, Iveraldo Ferreira da Silva, Maxwell Furtado de Lima, Assis dos Prazeres, and Luiz Roberto Oliveira da Costa for technical support during viral isolation and serologic tests.
This study was supported by Instituto Evandro Chagas/Secretária de Vigiláncia em Saúde/Ministry of Health, Salobo Metais S/A, and CNPq (process 300460/2005-8).
References
- Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. Virus taxonomy: classification and nomenclature of viruses. Eighth report of the International Committee on the Taxonomy of Viruses. San Diego (CA): Academic Press; 2005.
- Pinheiro FP, Travassos da Rosa AP, Travassos da Rosa JF, Ishak R, Freitas RB, Gomes ML, Oropouche virus. I. A review of clinical, epidemiological, and ecological findings. Am J Trop Med Hyg. 1981;30:149–60.PubMedGoogle Scholar
- Pinheiro FP, Travassos da Rosa AP, Vasconcelos PF. Oropouche fever. In: Feigin RD, editor. Textbook of pediatric infectious diseases, 5th ed. Philadelphia: Saunders; 2004. p. 2418–23.
- Pinheiro FP, Pinheiro M, Bensabath G, Causey OR, Shope RE. Epidemia de vírus Oropouche em Belém. Revista do Servico Especial Saude Publica. 1962;12:15–23.
- Saeed MF, Wang H, Nunes MRT, Vasconcelos PFC, Weaver SC, Shope RE, Nucleotide sequences and phylogeny of the nuclecapsid gene of Oropouche virus. J Gen Virol. 2000;81:743–8.PubMedGoogle Scholar
- Nunes MR, Martins LC, Rodrigues SG, Chiang JO, Azevedo RS, Travassos da Rosa AP, Oropouche virus isolation, southeast Brazil. Emerg Infect Dis. 2005;11:1610–3.PubMedGoogle Scholar
- Clarke DH, Casals J. Techniques for hemagglutination and hemagglutination-inhibition with arthropod-borne viruses. Am J Trop Med Hyg. 1958;7:561–73.PubMedGoogle Scholar
- Vasconcelos PF, Travassos da Rosa JF, Guerreiro SC, Dégallier N, Travassos da Rosa ES, Travassos da Rosa AP. Primeiro registro de epidemias causadas pelo vírus Oropouche nos estados do Maranhão e Goiás, Brasil. Rev Inst Med Trop Sao Paulo. 1989;31:271–8.PubMedGoogle Scholar
- Beaty BJ, Calisher CH, Shope RE. Arboviruses. In: Lennette DA, Schmidt NJ, editors. Diagnostic procedures for viral, rickettsial and chlamydial infections. Washington: American Public Health Association; 1995. p. 797–855.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstruction phylogenetic trees. Mol Biol Evol. 1987;4:406–25.PubMedGoogle Scholar
- Kumar S, Tamura K, Nei M. Molecular evolutionary genetic analysis. version 1.01. University Park (PA): The Pennsylvania State University; 2000.
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91. DOIGoogle Scholar
- Azevedo RS, Souza MR, Rodrigues SG, Nunes MR, Buna BS, Leão RNQ, Ocorrência endêmica de febre por Oropouche em Belém/PA no período de 2000 a 2001. Rev Soc Bras Med Trop. 2002;35(Suppl I):386.
Figures
Tables
Cite This ArticleTable of Contents – Volume 13, Number 6—June 2007
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Pedro Fernando da Costa Vasconcelos, Seção de Arbovirologia e Febres Hemorrágicas do Instituto Evandro Chagas/SVS/MS, Ave Almirante Barroso, 492, CEP 66093-020, Belém, Pará, Brazil;
Top