Volume 14, Number 7—July 2008
Peer Reviewed Report Available Online Only
Toward a Unified Nomenclature System for Highly Pathogenic Avian Influenza Virus (H5N1)
Figure 1
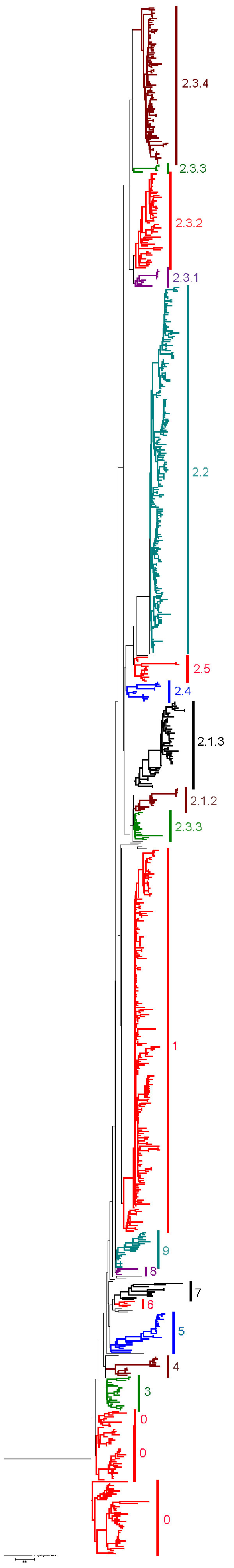
Figure 1. . . Neighbor-joining tree of 859 H5N1 isolates constructed by using PAUP* version 4.0b10 (9) with 1,000 bootstrap replicates. The tree was rooted by using the highly pathogenic avian influenza virus (H5N1) strain A/turkey/England/50–92/91, a historical European H5N1 virus closely related to the Gs/GD lineage (10). Clades are color coded with isolates not given a clade designation in light green. Maximum-likelihood trees used for comparison were constructed by using GARLI version 0.951 (11). Bayesian analysis for comparison was conducted with MrBayes version 3.1 (12) by using 4 replicates of 2 million generations, sampled every 100 generations, with 6 chains. The convergence of Markov Chain Monte Carlo chains was confirmed for each dataset by using Tracer version 1.3 (http://tree.bio.ed.ac.uk/software/tracer). Estimates of the statistical significance of phylogenies were calculated by performing 1,000 NJ bootstrap replicates, and Bayesian posterior probabilities were calculated from the consensus of 60,000 trees after excluding the first 20,000 trees (25%) as burn-in. Scale bar represents 0.01-nt changes.
References
- Duan L, Campitelli L, Fan XH, Leung YH, Vijaykrishna D, Zhang JX, Characterization of low-pathogenic H5 subtype influenza viruses from Eurasia: implications for the origin of highly pathogenic H5N1 viruses. J Virol. 2007;81:7529–39. DOIPubMedGoogle Scholar
- Ducatez MF, Olinger CM, Owoade AA, Tarnagda Z, Tahita MC, Sow A, Molecular and antigenic evolution and geographical spread of H5N1 highly pathogenic avian influenza viruses in western Africa. J Gen Virol. 2007;88:2297–306. DOIPubMedGoogle Scholar
- Chen H, Smith GJD, Li KS, Wang J, Fan XH, Rayner JM, Establishment of multiple sublineages of H5N1 influenza virus in Asia: Implications for pandemic control. Proc Natl Acad Sci U S A. 2006;103:2845–50. DOIPubMedGoogle Scholar
- Salzberg SL, Kingsford C, Cattoli G, Spiro DJ, Janies DA, Aly MM, Genome analysis linking recent European and African influenza (H5N1) viruses. Emerg Infect Dis. 2007;13:713–8.PubMedGoogle Scholar
- Smith GJD, Fan XH, Wang J, Li KS, Qin K, Zhang JX, Emergence and predominance of an H5N1 influenza variant in China. Proc Natl Acad Sci U S A. 2006;103:16936–41. DOIPubMedGoogle Scholar
- Smith GJD, Naipospos TSP, Nguyen TD, de Jong MD, Vijaykrishna D, Usman TB, Evolution and adaptation of H5N1 influenza virus in avian and human hosts in Indonesia and Vietnam. Virology. 2006;350:258–68. DOIPubMedGoogle Scholar
- Wallace RG, Hodac H, Lathrop RH, Fitch WM. A statistical phylogeography of influenza A H5N1. Proc Natl Acad Sci U S A. 2007;104:4473–8. DOIPubMedGoogle Scholar
- Nylander JAA. MRMODELTEST 2. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden; 2004 [cited 2007 Mar 12]. Available from http://www.abc.se/~nylander
- Swofford DL. PAUP: phylogenetic analysis using parsimony, version 4. Sunderland (MA): Sinauer Academic Publishers; 2001.
- Wood GW, Banks J, McCauley JW, Alexander DJ. Deduced amino acid sequences of the haemagglutinin of H5N1 avian influenza virus isolates from an outbreak in turkeys in Norfolk, England. Arch Virol. 1994;134:185–94. DOIPubMedGoogle Scholar
- Zwickl D. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD thesis, University of Texas at Austin; 2006 [cited 2008 Jun 5]. Available from http://www.bio.utexas.edu/faculty/antisense/garli/Garli.html
- Huelsenbeck JP, Ronquist FR. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5. DOIPubMedGoogle Scholar
- Kumar S, Tamura K, Nei M. MEGA3: an integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. DOIPubMedGoogle Scholar
- Fitch WM, Leiter JM, Li XQ, Palese P. Positive Darwinian evolution in human influenza A viruses. Proc Natl Acad Sci U S A. 1991;88:4270–4. DOIPubMedGoogle Scholar
- Wolf YI, Viboud C, Holmes EC, Koonin EV, Lipman DJ. Long intervals of stasis punctuated by bursts of positive selection in the seasonal evolution of influenza A virus. Biol Direct. 2006;1:34. DOIPubMedGoogle Scholar
1The working group was established by request of the World Health Organization’s Global Influenza Programme, Department of Epidemic and Pandemic Alert and Response (WHO, GIP, EPR), the World Organisation for Animal Health (OIE), and the Food and Agriculture Organization (FAO). It consisted of the following members: Ruben O. Donis, Centers for Disease Control and Prevention (CDC), Atlanta, Georgia, USA (co-chair); Gavin J.D. Smith, University of Hong Kong, Hong Kong Special Administrative Region, People’s Republic of China (co-chair); Michael L. Perdue, WHO, GIP, EPR, Geneva, Switzerland2 (coordinator); Ian H. Brown, Veterinary Laboratories Agency, Addlestone, UK; Hualan Chen, Harbin Veterinary Research Institute, Chinese Academy of Agriculture Sciences CAAS, Harbin, People’s Republic of China; Ron A.M. Fouchier, Erasmus University, Rotterdam, the Netherlands; Yoshihiro Kawaoka, University of Wisconsin-Madison, Madison, Wisconsin, USA, and Institute of Medical Science, University of Tokyo, Tokyo, Japan; John Mackenzie, Curtin University of Technology, Perth, Western Australia, Australia; and Yuelong Shu, China Centers for Disease Control, Bejing, People’s Republic of China. In addition, the following persons made substantial contributions: Ilaria Capua, Instituto Zooprofilattico Sperimentale delle Venezie, Padova, Italy; Nancy Cox, Todd Davis, Rebecca Garten, and Catherine Smith, CDC; Yi Guan and Dhanasekaran Vijaykrishna, University of Hong Kong; Elizabeth Mumford, WHO, GIP, EPR; and Colin A. Russell and Derek Smith, University of Cambridge, Cambridge, UK.
2Current affiliation: US Department of Health and Human Services, Washington, DC, USA.