Volume 15, Number 1—January 2009
Dispatch
Clonal Multidrug-Resistant Corynebacterium striatum Strains, Italy
Cite This Article
Citation for Media
Abstract
We assessed the clinical relevance and performed molecular characterization of 36 multidrug-resistant strains of Corynebacterium striatum. Pulsed-field gel electrophoresis confirmed a single clone, possessing erm(X), tetA/B, cmxA/B, and aphA1 genes, but few related subclones. This strain is emerging as a pathogen in Italy.
Isolation of Corynebacterium spp. as the only organism from clinical specimens from patients, mostly with varying degrees of immunocompromisation and severe infections, is increasing in Italy. Therefore, we evaluated the microbiologic characteristics, resistance profiles, and similarities among genomes of multidrug-resistant (MDR) C. striatum strains.
We evaluated 36 strains of MDR C. striatum, isolated from 3 hospitals in Italy during 2005–2007. Fourteen strains were from bronchoalveolar lavage (BAL) fluid, 3 from blood, 7 from central venous catheter tips, 5 from tracheal aspirates, 4 from wound specimens, 1 from BAL and pleural fluid, 1 from urine, and 1 from a lung biopsy specimen. To assess the clinical relevance of these strains, we used the Centers for Disease Control and Prevention 2004 definition for nosocomial infections (www.cdc.gov/ncidod/dhqp/nnis_pubs.html) (1) and tracked antimicrobial drug–resistance determinants.
We identified all strains as putative C. striatum by using the commercial system API 20 Coryne (bioMérieux, Marcy l’Etoile, France). C. striatum was differentiated from C. amycolatum by supplementary tests, i.e., tyrosine hydrolysis, N-acetylglucosamine assimilation, and phenylacetic acid assimilation (2); it was reconfirmed by sequencing the internal fragment of the 16S rRNA gene (3). The American Type Culture Collection (ATCC) 6940 C. striatum strain was included as phenotypic and molecular control. All strains were stored at –80°C until use.
MICs were determined by using microdilution in cation-adjusted Mueller-Hinton broth in accordance with guidelines of the Clinical and Laboratory Standards Institute (CLSI) (4). The following antimicrobial drugs were tested: tigecycline and piperacillin/tazobactam, oxacillin, gentamicin, kanamycin, levofloxacin, erythromycin, clindamycin, piperacillin, vancomycin, teicoplanin, tetracycline, moxifloxacin, imipenem, meropenem, quinupristin/dalfopristin, linezolid, and daptomycin. Etest strips (AB-BIODISK, Solna, Sweden) were used for vancomycin, teicoplanin, linezolid, and daptomycin. Daptomycin Etests were performed by using Muller-Hinton agar (Oxoid, Milan, Italy), supplemented to a final concentration of 50 mg/L calcium.
In the absence of approved breakpoints for Corynebacterium spp., we used those for α-hemolytic streptococci of the viridans group. Results were read after incubation at 37°C for 18–24 h. Susceptibility to daptomycin was defined as MIC <1 mg/L (5); CLSI guideline MIC breakpoints were used for all other drugs tested (4).
To further characterize the C. striatum isolates, we used 2 DNA fingerprinting techniques: automated ribotyping (RiboPrinter Microbial Characterization System; DuPont Qualicon, Wilmington, DE, USA) with EcoRI as restriction enzyme and pulsed-field gel electrophoresis (PFGE) macrorestriction analysis with 2 enzymes (XbaI and SwaI; New England Biolabs, Beverly, MA, USA). We had used 4 enzymes (XbaI, SwaI, SfiI, and PacI) to test 10 random strains, but because XbaI and SwaI enzyme-restriction patterns gave a better resolution for low and high molecular weight fragments, respectively, we used only these 2 restriction enzymes to type all 36 strains.
Whole genomic DNA chromosomal extraction, macrorestriction digestion, and PFGE (CHEF-DR II apparatus; Bio-Rad, Hercules, CA, USA) were performed as previously reported (6). Macrorestriction fragments were separated on 1% (wt/vol) ultrapure agarose gels (Sigma Aldrich, St. Louis, MO, USA) at 6 V/cm, for 21 h at 14°C with pulse times of 0.1–5 s, to separate XbaI fragments, and for 23 h with pulse times of 1–70 s, to separate SwaI fragments. Lambda DNA concatemers (New England BioLabs) were used as molecular size markers. Similarities among macrorestriction patterns were identified according to established criteria (7).
The sequence of pTP10 (GenBank accession no. AF024666) (8) was used to design the primers for erm(X), tetA and tetB, cmx, aphA1, and repB genes. The VectorNTI program (Invitrogen, www.invitrogen.com) was used for this purpose. The presence of pTP10 was confirmed first by amplification and sequencing of the resistance determinants and the replication gene (repB) and then by XbaI and SwaI PFGE hybridizations, performed with the specific probes (erm(X), tetAB, cmx, and aphA1), following a protocol previously described (9). The PCR amplifications were performed in a Techne TC412 thermal cycler (Barloworld Scientific, Staffordshire, UK). All primers and the related probe regions used in hybridization experiments are shown in Table 1.
All C. striatum isolates were recovered from hospitalized patients who had undergone surgery or been admitted to intensive care units (Table 2). We documented 19 cases of infections and discarded 17 as contaminants. The isolates that were considered causes of infections were responsible for 8 cases of ventilator-associated pneumonia (including 1 with associated pleural empyema), 2 cases of pneumonia, 1 case of catheter-related sepsis, 2 cases of ventilator-associated tracheobronchitis, and 6 cases of wound infections.
The 36 strains showed an MDR phenotype, including resistance to >3 classes of drugs; MICs required to inhibit growth of 90% (MIC90) were penicillins >256 mg/L, carbapenems >256 mg/L, gentamicin 32 mg/L, levofloxacin 256 mg/L, tetracycline >256 mg/L, lincosamides >256 mg/L, and erythromycin 32 mg/L. C. striatum strains were susceptible to only the most recent drugs used for treatment of infections with gram-positive organisms, such as glycopeptides and tigecycline (MIC90 1 mg/L), quinupristin/dalfopristin and daptomycin (MIC90 0.25 mg/L), and linezolid (MIC90 2 mg/L). A discrepancy was found when susceptibility testing using a disk-diffusion method was performed on different strains; the inhibition zone of erythromycin was always in the intermediate range, even if MICs for this drug were in the low-resistance range.
Ribotyping gave a unique profile for all strains in this study. PFGE enabled us to discriminate the right number of macrorestriction fragments (5,10,11) for pattern comparison.
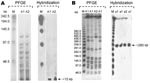
Analyses of SwaI digestion patterns showed that of the 36 strains, only 1 clone had 3 different subtypes (30 strains subtype a1, 4 strains a2, and 2 strains a3). Macrorestriction analysis with XbaI showed almost comparable results (27 strains A1, 7 strains A2, and 2 strains A3) (Figure). This genotyping method and the enzymes used were defined as appropriate, comparing PFGE patterns of our clinical isolates with C. striatum ATCC 6940 type strain, which was different with respect to the epidemic strains. This result demonstrates that single MDR C. striatum clones had been selected and were circulating in the 3 hospitals.
Further, the molecular characterization of some of the resistance genes in the 36 C. striatum isolates demonstrated the presence of erm(X), codifying for the resistance to erythromycin and clindamycin; tetA, and tetB, codifying for the resistance to tetracycline, oxytetracycline, and oxacillin; and cmx and aphA1, responsible for resistance to aminoglycosides and chloramphenicol, respectively. The presence of pTP10 carrying all these determinants was confirmed by amplification and sequencing of these genes and the replication gene of the plasmid, together with hybridization experiments demonstrating that all resistance determinants were localized in the same hybridization band generated by each probe onto PFGEXbaI (≈15 kb) and PFGESwaI (≈280 kb) membranes (Figure).
We report isolation of MDR C. striatum from clinical specimens responsible for cases of pneumonia, catheter-related bacteremia, and wound infections. Infections sustained from this species are strongly associated with devices, not only tubes or catheters (91%) but also sternal surgical wound wires.
The MDR phenotype of these strains was immediately observed and was responsible for the alarm that led to the subsequent in-depth examination of these strains. Their clonal nature, as demonstrated in our study, is of particular concern. Further, the MDR phenotype correlated to the presence of the pTP10 plasmid, which demonstrates that these MDR microorganisms acquired not only the capability to cause infections but also increased resistance and the ability to spread by virtue of their clonal nature. The only drugs still active against these MDR strains are glycopeptides, linezolid, quinopristin/dalfopristin, daptomycin, and tigecycline. To avoid using drugs that appear active in vitro but that could be ineffective in vivo, clinicians should be aware of the circulation of these MDR strains.
Dr Campanile is a researcher at the Department of Microbiology, University of Catania. She is involved in the fields of antimicrobial drug resistance, molecular typing, evolutionary relationships among strains of diverse sources, and horizontal exchange of antimicrobial drug resistance determinants by mobile genetic elements.
Acknowledgments
We are indebted to Antony Brigdewood for the language revision of the manuscript.
This work was supported by grants from European Union Drug Resistance Spread 2 project contract no. 018705 to S.S. and from Fondazione IRCCS Policlinico San Matteo, Pavia, to E.C. (Ricerca Finalizzata 2006) and to P.M. (Ricerca Corrente 1998–2006: “Sorveglianza delle infezioni ospedaliere: tipizzazione epidemiologica di microrganismi patogeni mediante metodiche molecolari”).
References
- Mayall C, ed. Surveillance of nosocomial infections. 3rd ed. Philadelphia: Lippincott Williams & Wilkins; 2004.
- Renom F, Garau M, Rubi M, Ramis F, Galmes A, Soriano JB. Nosocomial outbreak of Corynebacterium striatum infection in patients with chronic obstructive pulmonary disease. J Clin Microbiol. 2007;45:2064–7. DOIPubMedGoogle Scholar
- Pascual C, Lawson PA, Farrow JA, Gimenez MN, Collins MD. Phylogenetic analysis of the genus Corynebacterium based on 16S rRNA gene sequences. Int J Syst Bacteriol. 1995;45:724–8.PubMedGoogle Scholar
- Clinical and Laboratory Standard Institute. Performance standards for antimicrobial testing. Approved standards. Wayne (PA): The Institute; 2006.
- Iaria C, Stassi G, Costa GB, Biondo C, Gerace E, Noto A, Outbreak of multi-resistant Corynebacterium striatum infection in an Italian general intensive care unit. J Hosp Infect. 2007;67:102–4. DOIPubMedGoogle Scholar
- Sampaio JLM, Chimara E, Ferrazoli L, da Silva Telles MA, Del Guercio VM, Jerico ZVN, Application of four molecular typing methods for analysis of Mycobacterium fortuitum group strains causing post-mammaplasty infections. Clin Microbiol Infect. 2006;12:142–9. DOIPubMedGoogle Scholar
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9.PubMedGoogle Scholar
- Tauch A, Krieft S, Kalinowski J, Puhler A. The 51,409-bp R-plasmid pTP10 from the multiresistant clinical isolate Corynebacterium striatum M82B is composed of DNA segments initially identified in soil bacteria and in plant, animal, and human pathogens. Mol Gen Genet. 2000;263:1–11. DOIPubMedGoogle Scholar
- Mato R, Camapnile F, Stefani S, Crisostomo MI, Santagati M, Sanches SI, Clonal types and multidrug resistance patterns of methicillin-resistant Staphylococcus aureus (MRSA) recovered in Italy during the 1990s. Microb Drug Resist. 2004;10:106–13. DOIPubMedGoogle Scholar
- Tarr PE, Stock F, Cooke RH, Fedorko DP, Lucey DR. Multidrug-resistant Corynebacterium striatum pneumonia in a heart transplant recipient. Transpl Infect Dis. 2003;5:53–8.PubMedGoogle Scholar
- Martin MC, Melon O, Celada MM, Alvarez J, Mendez FJ, Vazquez F. Septicaemia due to Corynebacterium striatum: molecular confirmation of entry via the skin. J Med Microbiol. 2003;52:599–602. DOIPubMedGoogle Scholar
Figure
Tables
Cite This ArticleTable of Contents – Volume 15, Number 1—January 2009
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Stefania Stefani, Department of Microbiology, University of Catania, Via Androne 81, 95124 Catania, Italy;
Top