Volume 15, Number 2—February 2009
Dispatch
Novel Human Parechovirus from Brazil
Figure 1
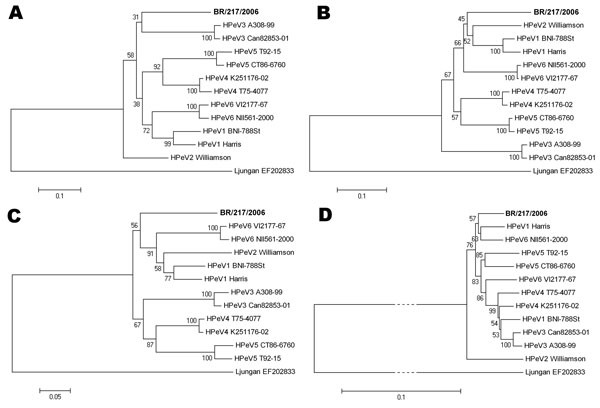
Figure 1. Evolutionary relationships between known parechoviruses and the new human parechovirus from this study (boldface). Phylogenetic analyses of A) complete viral protein (VP) 1, B) VP0, and C) VP3, and D) the whole nonstructural region (comprising regions P2 and P3), were constructed using the Jones-Taylor-Thornton matrix-based substitution model on an amino acid–driven alignment (pairwise deletion option). The evolutionary histories were inferred using the neighbor-joining method and are in the units of the number of amino acid substitutions per site. Relevant bootstrap values from 500 replicate trees are shown next to the branches. The scale bars show the evolutionary distance from each root. Analysis was conducted in MEGA software version 4 (www.megasoftware.net). HPeV reference strains are named in full detail; their GenBank accession numbers are not further indicated. Contemporary HPeV1 strain BNI-788st accession number was EF051629 and HPeV6 strain BNI-67 accession number was EU022171. The tree was rooted against Ljungan virus, a rodent parechovirus (GenBank accession no. EF202833). In the P2/P3 tree, branches to the Ljungan virus node have been truncated for space reasons, as indicated by dotted lines.