Volume 15, Number 2—February 2009
Dispatch
Novel Human Parechovirus from Brazil
Figure 2
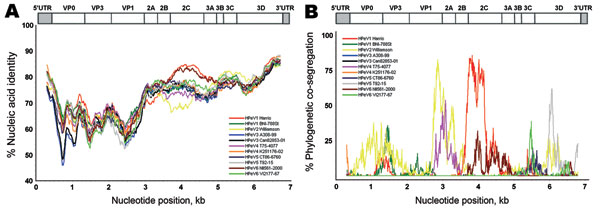
Figure 2. Nucleic acid identity with known parechoviruses. The near full-length genome of the new parechovirus BR/217/2006 was analyzed with SimPlot software (http://sray.med.som.jhmi.edu/SCRoftware/simplot) using a 600-bp sliding window and a step size of 10. Because of partially incomplete 5′ untranslated (UTR) region GenBank reference sequences, an approximate 400 nucleotides had to be cut from the 5′ end of all genomes. HPeV, human parechovirus. A) Nucleic acid identity, per analysis window, for strain BR/217/2006 with prototype strains. Nucleotide positions on x-axis show the center of the window. B) BootScan analysis using the same window settings. A bootstrapped phylogenetic analysis was conducted per window along the alignment. Graphs represent the percentage (bootstrap values) at which each strain cosegregates phylogenetically in the analysis window with strain BR/217/2006. Prototype strains used for comparison are shown in the inserts in each panel. A schematic representation of the parechovirus genome is given on top.