Detection of Novel SARS-like and Other Coronaviruses in Bats from Kenya
Suxiang Tong

, Christina Conrardy, Susan Ruone, Ivan V. Kuzmin, Xiling Guo, Ying Tao, Michael Niezgoda, Lia M. Haynes, Bernard Agwanda, Robert F. Breiman, Larry J. Anderson, and Charles E. Rupprecht
Author affiliations: Centers for Disease Control and Prevention, Atlanta, Georgia, USA (S. Tong, C. Conrardy, S. Ruone, I.V. Kuzmin, Y. Tao, M. Niezagoda, L. Haynes, L.J. Anderson, C.E. Rupprecht); Jiangsu Provincial Center for Disease Control and Prevention, Nanjing, People’s Republic of China (X. Guo); National Museum, Kenya Wildlife Service, Nairobi, Kenya (B. Agwanda); Centers for Disease Control and Prevention Kenya, Nairobi (R.F. Briman)
Main Article
Figure 2
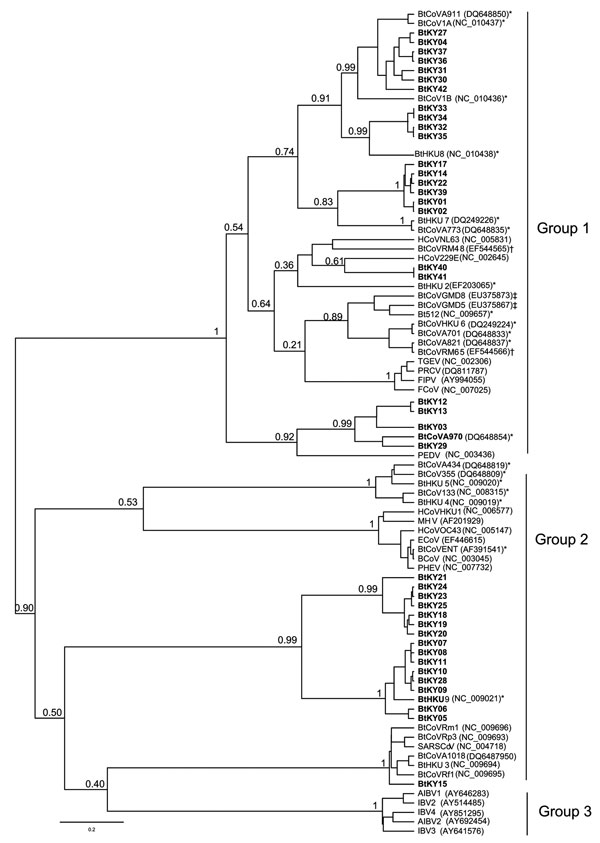
Figure 2. Phylogenetic tree generated using Bayesian Markov Chain Monte Carlo analysis implemented in Bayesian Evolutionary Analysis Sampling Trees (BEAST; http://beast.bio.ed.ac.uk) by using a 121-nt fragment of the RdRp gene 1b from 39 coronaviruses (CoVs) in bats from Kenya. CoVs from this study are shown in boldface; an additional 47 selected human and animal coronaviruses from the National Center for Biotechnology Information database are included. The Bayesian posterior probabilities were given for deeper nodes. CoV groups (1 to 3) based on International Committee on Taxonomy of Viruses recommendation are indicated. Bat coronaviruses from the People’s Republic of China (*), northern Germany (†), and North America (‡) are labeled. Scale bar indicates number of nucleotide substitutions per site.
Main Article
Page created: December 07, 2010
Page updated: December 07, 2010
Page reviewed: December 07, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
