Volume 15, Number 5—May 2009
Dispatch
Possible Seasonality of Clostridium difficile in Retail Meat, Canada
Figure 2
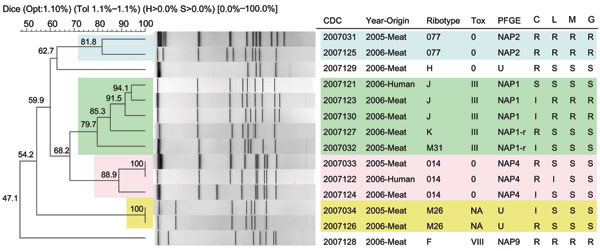
Figure 2. Pulsed-field gel electrophoresis (PFGE)–SmaI dendogram of Clostridium difficile isolates of meat and human origin in Canada. Representative PCR ribotypes 077, 014, M31, and M26 are of meat origin from 2005 (4,11). PCR ribotype designations are described in Table 2. Note the genetic similarity (94.1%–100%) and antimicrobial resistance profiles between human and meat isolates, especially PCR ribotypes 014 and J. Also note the genetic similarity (81.8%–100%) between meat isolates from 2005 and 2006 for multidrug-resistant epidemic PCR ribotype 077, clindamycin-variable, PCR ribotype 014, and nontoxigenic PCR ribotype M26. Resistance to all 4 antimicrobial drugs was observed in meat isolates of ribotypes 077 and F, which also yielded the highest level of clindamycin resistance (>256 µL/mL; breakpoint: >6 µL/mL). The breakpoints for moxifloxacin (12) were also used for levofloxacin and gatifloxacin. R (resistant), S (susceptible), and I (intermediate) represent antimicrobial profiles. CDC, Centers for Disease Control and Prevention; NAP, North America PFGE type; NAP1-r, NAP-related strain; Tox, toxinotyping nomenclature (M. Rupnik, Maribor, Slovenia); U, unnamed.
References
- McDonald LC, Killgore GE, Thompson A, Owens RC Jr, Kazakova SV, Sambol JP, An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433–41. DOIPubMedGoogle Scholar
- Martin H, Willey B, Low DE, Staempfli HR, McGeer A, Boerlin P, Characterization of Clostridium difficile isolated from patients in Ontario, Canada, from 2004 to 2006. J Clin Microbiol. 2008;46:2999–3004. DOIPubMedGoogle Scholar
- Barbut F, Mastrantonio P, Delmee M, Brazier J, Kuijper E, Poxton I. European Study Group on Clostridium difficile (ESGCD). Prospective study of Clostridium difficile infections in Europe with phenotypic and genotypic characterisation of the isolates. Clin Microbiol Infect. 2007;13:1048–57. DOIPubMedGoogle Scholar
- Rodriguez-Palacios A, Staempfli HR, Duffield T, Weese JS. Clostridium difficile in retail ground meat, Canada. Emerg Infect Dis. 2007;13:485–7.PubMedGoogle Scholar
- Public Health Agency of Canada. Canadian integrated program for antimicrobial resistance surveillance (CIPARS) 2005 [cited 2008 Jan 20]. Available from http://www.phac-aspc.gc.ca/cipars-picra/index-eng.php
- Arroyo LG, Rousseau J, Willey BM, Low DE, Staempfli H, McGeer A, Use of a selective enrichment broth to recover Clostridium difficile from stool swabs stored under different conditions. J Clin Microbiol. 2005;43:5341–3. DOIPubMedGoogle Scholar
- Lemee L, Dhalluin A, Testelin S, Mattrat MA, Maillard K, Lemeland JF, Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (toxin A), and tcdB (toxin B) genes for toxigenic culture of Clostridium difficile. J Clin Microbiol. 2004;42:5710–4. DOIPubMedGoogle Scholar
- Spigaglia P, Mastrantonio P. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J Clin Microbiol. 2002;40:3470–5. DOIPubMedGoogle Scholar
- Bidet P, Barbut F, Lalande V, Burghoffer B, Petit JC. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett. 1999;175:261–6. DOIPubMedGoogle Scholar
- Rupnik M, Dupuy B, Fairweather NF, Gerding DN, Johnson S, Just I, Revised nomenclature of Clostridium difficile toxins and associated genes. J Med Microbiol. 2005;54:113–7. DOIPubMedGoogle Scholar
- Rodriguez-Palacios A, Stampfli HR, Duffield T, Peregrine AS, Trotz-Williams LA, Arroyo LG, Clostridium difficile PCR ribotypes in calves, Canada. Emerg Infect Dis. 2006;12:1730–6.PubMedGoogle Scholar
- Clinical and Laboratory Standards Institute (CLSI). Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard, seventh edition. CLSI document M11–A7; 2007:27(2) [cited 2008 May 17]. Available from http://www.clsi.org/source/orders/free/m11a7.pdf
- Songer JG, Trinh HT, Thompson AD, Killgore GE, McDonald L, Limbago BM. Isolation of Clostridium difficile from retail meats. In: Second International Clostridium difficile Symposium; June 6–9, 2007; Maribor, Slovenia. Maribor (Slovenia): Marie Curie Conferences and Training Procedures; 2007. p. 44.
- Vengust M, Arroyo LG, Weese JS, Baird JD. Preliminary evidence for dormant clostridial spores in equine skeletal muscle. Equine Vet J. 2003;35:514–6. DOIPubMedGoogle Scholar
- National Health Service for Wales. All-Wales mandatory Clostridium difficile surveillance- 2006 report [cited 2007 Jun 21]. Available from http://www.wales.nhs.uk/sites3/page.cfm?orgid=379&pid=18490