Volume 15, Number 7—July 2009
Dispatch
Genetically Diverse Coronaviruses in Wild Bird Populations of Northern England
Appendix Figure
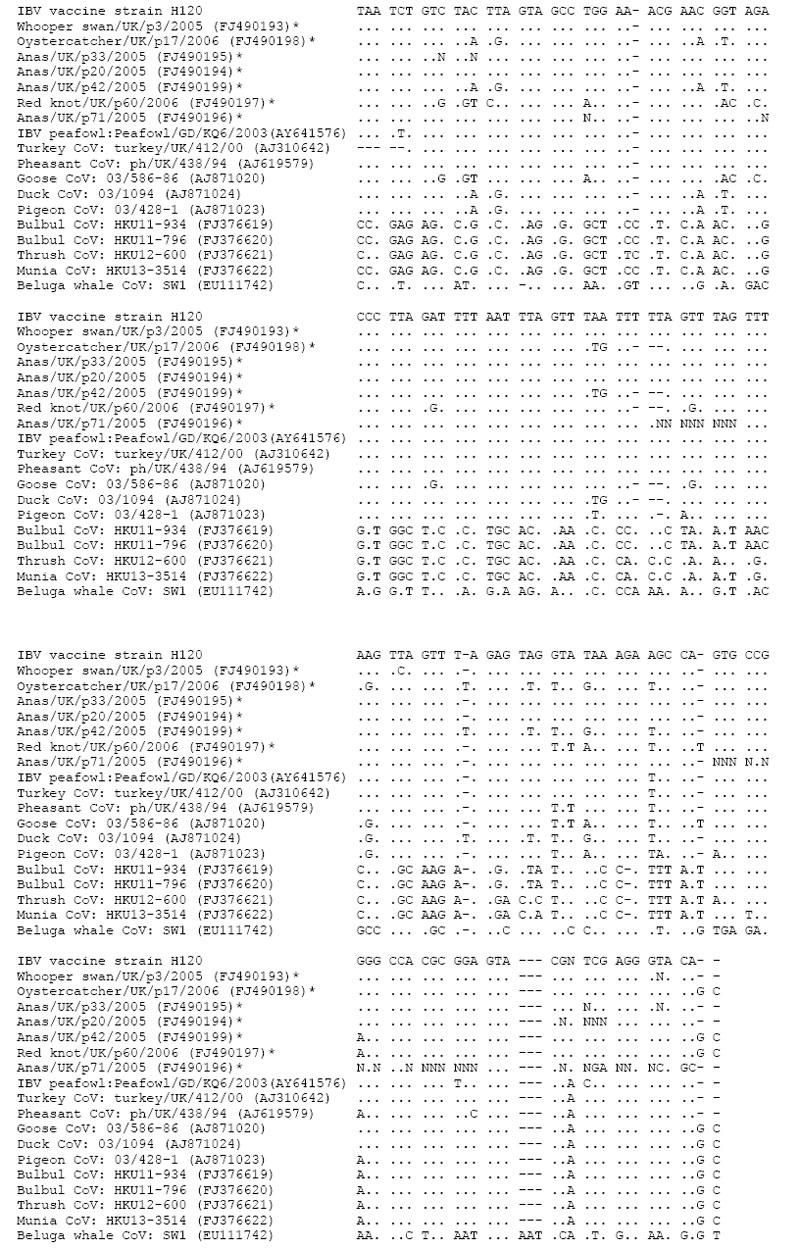
Appendix Figure. Multiple-sequence alignment of a fragment of the 3? untranslated region of coronaviruses detected in wild birds in this study and other previously published group 3 coronavirus sequences from wild birds and a beluga whale. Viruses detected by this study are marked with an asterisk. GenBank accession numbers for all sequences are shown in parentheses. Identical nucleotides are marked with a period (.). Sequences were aligned using the Clustal program within the MEGA 4.0 software package (10).
Page created: November 09, 2010
Page updated: November 09, 2010
Page reviewed: November 09, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.