Volume 17, Number 2—February 2011
Dispatch
Usefulness of Published PCR Primers in Detecting Human Rhinovirus Infection
Figure
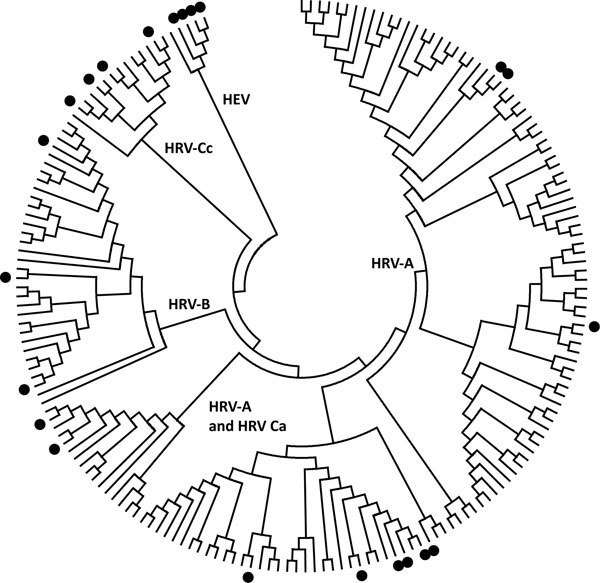
Figure. Distribution of human rhinovirus (HRV) and human enterovirus (HEV) sequences used for primer pair studies. The HRV and HEV genotypes from the testing panel (indicated by filled circles) were aligned with the central 154 nt of the 5′ untranslated region (UTR) region of all complete HRV genomes and poliovirus-1. HRV-Ca and HRV-Cc refer to HRV-Cs with 5′ UTR sequences that have phylogenetic origins from either HRV-As or HRV-Cs, respectively. The tree was constructed by neighbor joining of maximum composite likelihood distance implemented in MEGA (www.megasoftware.net).
Page created: July 13, 2011
Page updated: July 13, 2011
Page reviewed: July 13, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.