Volume 17, Number 5—May 2011
Research
Genotypic Profile of Streptococcus suis Serotype 2 and Clinical Features of Infection in Humans, Thailand
Cite This Article
Citation for Media
Abstract
To examine associations between clinical features of Streptococcus suis serotype 2 infections in humans in Thailand and genotypic profiles of isolates, we conducted a retrospective study during 2006–2008. Of 165 patients for whom bacterial cultures of blood, cerebrospinal fluid, or both were positive for S. suis serotype 2, the major multilocus sequence types (STs) found were ST1 (62.4%) and ST104 (25.5%); the latter is unique to Thailand. Clinical features were examined for 158 patients. Infections were sporadic; case-fatality rate for adults was 9.5%, primarily in northern Thailand. Disease incidence peaked during the rainy season. Disease was classified as meningitis (58.9%) or nonmeningitis (41.1%, and included sepsis [35.4%] and others [5.7%]). Although ST1 strains were significantly associated with the meningitis category (p<0.0001), ST104 strains were significantly associated with the nonmeningitis category (p<0.0001). The ST1 and ST104 strains are capable of causing sepsis, but only the ST1 strains commonly cause meningitis.
Streptococcus suis, an emerging zoonotic pathogen, causes invasive infections in persons who are in close contact with infected pigs or contaminated pork-derived products (1). On the basis of capsular polysaccharides, 33 serotypes of S. suis have now been identified. Of these, serotype 2 is the most prevalent type in humans infected with this pathogen (1,2). Since the largest outbreak of human S. suis infection in 2005, in Sichuan Province, People’s Republic of China (3), this disease has been increasingly recognized worldwide. The numbers of reported cases, especially in persons from Southeast Asian countries, have increased dramatically during past few years (4).
In Thailand, at least 300 cases of S. suis infection in humans have been reported (5–11). Although an outbreak of S. suis infections was confirmed in Phayao Province during May 2007 (9), most cases in humans occur sporadically and are primarily located in the northern region of this country (6–11). A relatively low incidence of cases with S. suis serotype 14 has also been reported in this region (12). Although previous studies have reported high frequencies (59.0%–88.7%) of S. suis infections in persons in this area who ate raw pork products (8–11), the pathogenesis of this disease, including routes of transmission, is unclear.
The major clinical manifestations of the disease are bacterial meningitis and sepsis, but other manifestations have been reported (1,4,8,10,13). Most cases of bacterial meningitis can be attributed to the hematogenic spread of invasive bacteria, but how circulating bacteria cross the blood–cerebrospinal fluid (CSF) barrier and cause meningitis is not clear (14,15). Furthermore, the overall clinical features of this disease have not been extensively and comprehensively investigated in Southeast Asian countries.
A variety of virulence factors associated with S. suis have been reported (16–20), but none have been proven to be essential for the host defense of this disease, except the capsular polysaccharide (19). In serotype 2 isolates obtained during a previous outbreak in Sichuan, China, an ≈89-kb DNA fragment, which has been associated with a pathogenicity island (89K PAI), was identified (21). The 89K PAI fragment encodes a 2-compartment signal transduction system, SalK-SalR, which is required for full virulence (22).
We report the results of a retrospective study of the clinical features of 158 cases of human infection with S. suis serotype 2 and the molecular epidemiology of 165 S. suis serotype 2 isolates. The study objective was to demonstrate associations between the clinical features of disease caused by S. suis serotype 2 in persons in Thailand and the genotypic profiles of the isolates. The study was reviewed and approved by the Ethics Committees of Research Institute for Microbial Diseases, Osaka University, and conducted according to the principles expressed in the Declaration of Helsinki.
Isolate Identification
From January 2006 through August 2008, a total of 1,154 unidentified streptococcal isolates from blood or CSF were collected from hospitals in all 76 provinces of Thailand. Biochemical testing of these isolates, using API Strep (bioMérieux, Durham, NC, USA) and S. suis–specific and S. suis serotype 2– or 1/2–specific PCR (12,23), confirmed 165 isolates from 34 hospitals in 25 provinces as S. suis. The final serotype of all strains was confirmed by coagglutination tests that used rabbit antiserum (Statens Serum Institute, Copenhagen, Denmark).
Genotypic Profiles of Isolates
Multilocus sequence type (MLST) testing was performed as described by King et al. (24), with a modification for mutS as described by Rehm et al (25). MLST alleles and the resulting sequence type (ST) were assigned by using the S. suis MLST database (http://ssuis.mlst.net). eBURST was used to identify the clonal complexes for these 165 serotype 2 strains within S. suis, and the overall structure of the population was obtained through the MLST database (26). Virulence-associated genes (VAG), including extracellular released protein factor (epf), muramidase-released protein (mrp), and suilysin (sly), and variants of mrp or epf were determined by PCR as described by Silva et al. (27), with minor modifications. Presence of the 89K PAI fragment was determined by PCR as reported by Chen et al (27). Pulsed-field gel electrophoresis (PFGE) was performed as described (28), and the pulsotypes were assigned to clusters of isolates with >80% similarity from the dendrogram. The dendrogram representing the genetic relationships between the representative pulsotypes from 165 S. suis serotype 2 strains was drawn by using the Cluster 3.0 software program and examined by using the TreeView program as described (12,29).
Clinical Features of Cases
Of the 165 patients whose culture results were positive for S. suis serotype 2, medical records for 158 were retrospectively reviewed by physicians at local hospitals in Thailand. Medical records for the remaining 7 patients were not available. The clinical manifestations were mostly divided into 2 categories: meningitis and nonmeningitis. The meningitis category involved confirmed meningitis, bacteremic meningitis, and probable meningitis. All patients in the meningitis category had typical meningeal signs, such as neck stiffness, and acute disease onset. Although bacteremic meningitis was defined as a case in which both CSF and blood cultures were positive, confirmed meningitis was defined as a case with a positive CSF culture only, and probable meningitis was defined as a case with a positive blood culture only. The nonmeningitis category included the clinical manifestations of sepsis and sepsis with focal signs other than meningitis (septic arthritis or spondylodiscitis, infective endocarditis, and bacteremic pneumonia). Sepsis was defined as systemic inflammatory response syndrome and a positive blood culture (30), and septic arthritis or septic spondylodiscitis was defined as described (31). Diagnosis of infectious endocarditis was based on the Duke criteria (32). Septic shock was also defined as described (33).
Statistical Analyses
Comparisons of the clinical characteristics between fatal and nonfatal cases were analyzed by using the χ2 test or Fisher exact test with Stata version 10.0 software (StataCorp, College Station, TX, USA). Patient ages and periods of hospital admission were tested for normality of the distribution using the Kolmogorov-Smirnov test and were compared by using the Student t test with SPSS version 11.0 software (SPSS Inc., Chicago, IL, USA). Data were considered significant at p<0.05.
Genotypic Profiles of Isolates
Of the 165 S. suis serotype 2 isolates, 123 were isolated from blood and 42 from CSF. eBURST analysis based on MLST enabled classification of these strains into 4 ST complexes: the ST1, ST27, ST29, and ST104 complexes (Table 1). ST126, a novel ST, has a single locus variant from ST1. The largest cluster of 89K PAI–carrying strains was ST1 (n = 81, 49.1%), which had the epf+/sly+/mrp+ genotype; these strains were isolated from blood and CSF. Another large cluster of non-89K PAI–carrying strains was ST104, which had the epf–/sly+/mrp– genotype (n = 39, 23.6%); most of these strains (n = 38) were isolated only from blood. ST103, ST104, and ST126 were found only in isolates from humans in Thailand.
PFGE of Isolates
Of the 165 serotype 2 strains, PFGE analyses identified 20 pulsotypes (Figure 1, panel A). Analysis of the dendrogram for these 20 pulsotypes revealed at least 16 clusters (I to XVI) (Figure 1, panel B). Although 5 pulsotypes of A were identified for the ST1 and ST126 strains, 2 major pulsotypes (A [n = 32] and A1 [n = 43]), A1 (n = 43), and A4 (n = 3) were grouped in 1 cluster. Pulsotype A2 (n = 21), which consisted of ST1 strains lacking the 89K PAI fragment, was classified into a distinguished cluster. PFGE showed diverse DNA patterns for strains ST25 and ST103. ST25 strains were classified into 5 clusters of I, II, III, IV, and VIII. ST103 strains were classified into 3 clusters of VI, XIV, and XV. Three ST28 strains lacking 89K PAI exhibited the unique DNA pattern of pulsotype D; these were classified into cluster XVI. Although 4 pulsotypes (H, H1, H2, and H3) were identified for ST104 strains, 2 major pulsotypes (H [n = 29] and H1 [n = 11]) in ST104 strains were classified into cluster VII. Collectively, clusters X and XI for ST1 and ST126 strains and cluster VII for ST104 strains accounted for the major 3 clusters found for cases in Thailand.
Geographic and Seasonal Distribution
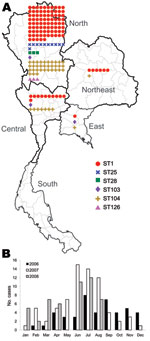
Of the 165 isolates, 136 (82.4%) were from the northern region, 19 (11.5%) from the central region, 7 (4.2%) from the northeast region, and 3 (1.8%) from the eastern region (Table 2; Figure 2, panel A). No strains were isolated from the southern region. The dates of isolation suggest that human cases occur more frequently during the rainy season, June–August of each year (Figure 2, panel B).
Clinical Features of Cases
The clinical features of the 158 human cases of S. suis serotype 2 infection are summarized in Table 3. The median age (range) of the 155 patients for whom age was known was 55.0 (18–93) years; 72.8% were male. No cases in children were identified in this study. All 158 patients had been hospitalized; median duration (range) of hospitalization for the 158 patients was 11 (1–45) days; 15 (9.5%) patients died. No significant differences were found between the fatal and nonfatal cases with respect to patient age or period of admission.
The meningitis category (n = 93) included 22 cases of confirmed meningitis, 44 cases of bacteremic meningitis, and 27 cases of probable meningitis. The nonmeningitis category (n = 65) included sepsis with focal signs other than meningitis (n = 9) and sepsis (n = 56). Sepsis with focal signs other than meningitis included septic arthritis (n = 5), infective endocarditis (n = 3), and bacteremic pneumonia (n = 1). Of the 15 fatal cases, 8 were assigned to the meningitis category (probable meningitis [n = 6], meningitis [n = 1], bacteremic meningitis [n = 1]), 6 cases were sepsis, and 1 case was infective endocarditis (Table 3). Although the cases of bacteremic meningitis were significantly associated with a nonfatal outcome (p = 0.043), the probable meningitis cases were significantly associated with a fatal outcome (p = 0.013). The combined frequencies for the recent consumption of raw pork products and exposure to pigs were 39.9%. None of the clinical signs or possible risk factors, including recent exposure to pigs or raw pork products, or alcohol abuse, was significantly associated with a fatal outcome. Of the 158 patients, 154 parenterally received antimicrobial drugs, such as ceftriaxone, and data concerning antimicrobial drug treatment were not available for 4. Corticosteroids, such as dexamethasone, were used for only 4 patients.
Clinical Features and Genotype Profiles
The distributions of STs for the 158 human isolates for the meningitis and nonmeningitis categories are shown in Table 4. Although the ST1 strains were significantly associated with the meningitis category (p<0.0001), the ST104 strains were significantly associated with the nonmeningitis category (p<0.0001). The VAG profile of epf+/sly+/mrp+, which was dominant in the ST1 strains, was also significantly associated with the meningitis category (p<0.0001). The VAG profile of epf–/sly+/mrp–, which was observed only in the ST104 strains, was also significantly associated with the nonmeningitis category (p<0.0001). Because the largest cluster of 89K PAI–carrying strains was associated with the VAG profile of epf+/sly+/mrp+, the presence of 89K PAI was also significantly associated with the meningitis category (p<0.0001). None of the genotypic profiles that included STs, VAG, and presence of 89K PAI were significantly associated with fatal or nonfatal outcomes (data not shown).
Our finding that isolated S. suis serotype 2 strains peaked during the rainy season of 2006–2008 confirmed conclusions reached in previous small-scale studies conducted in northern Vietnam and Hong Kong (35,36). The predominant distribution of these isolates in northern Thailand is also in accordance with previous reports (6–11). However, why no human cases were identified in southern Thailand remains uncertain. A recent study from Hong Kong reported heavy contamination of S. suis in raw pork meat at local supermarkets or wet markets; therefore, a hot and humid climate may facilitate the growth of S. suis in raw pork products in those markets (37) and increase the risk for S. suis infections in humans in northern Thailand. The finding of no cases in children suggests that the routes of transmission are associated with adult behavior.
A recent study from northern Thailand, based on 20 human isolates collected during 1998–2002, reported that the most common isolates of S. suis serotype 2 were ST25 (40%), followed by ST1 (15%) and ST103 (15%) (34). By contrast, the MLST and PFGE results in this study clearly demonstrated that ST1 strains with major pulsotypes of A, A1 and A2, and ST104 with major pulsotypes of H and H1 were currently circulating in the same region of Thailand during 2006–2008. Collectively, these data suggest dynamic replacement of STs from ST25 to ST1 and ST104 among serotype 2 strains during recent years in this region.
Although S. suis serotype 2 has been reported to be the most frequent cause of bacterial meningitis in adults in Vietnam (13,35), other clinical manifestations, such as sepsis and infectious endocarditis, have also been found to be common in Thailand (6,8,11). Of the 158 human cases in the study reported here, ≈60% were assigned to the meningitis category and ≈35% were sepsis. Other clinical manifestations, including infective endocarditis, were rare. The findings reported here demonstrate significant associations between the ST1 strains and the meningitis category and between the ST104 strains and the nonmeningitis category. These findings indicate that both the ST1 and ST104 strains cause bacteremia and sepsis but that the ST1 strains are more likely to cross the blood–CSF barrier and subsequently result in meningitis. Because ≈80% of the cases in the meningitis category were caused by strains with ST1, as evidenced by a VAG profile of epf+/sly+/mrp+ and 89K PAI, these genotypic profiles of S. suis serotype 2 may favor bacterial survival and multiplication in the bloodstream, which would result in high levels of bacteremia, crossing of the blood–CSF barrier, and invasion of the meninges and the central nervous system (15). Our PFGE data showed that the pulsotype A1 found in serotype 2 strains with ST1 was identical to pulsotype 11 of serotype 2 strains with ST1 from Vietnam and pulsotype I of the serotype 2 strains with ST1 from Hong Kong (13,28). These isolates from Vietnam and Hong Kong were associated with a VAG profile of epf+/sly+/mrp+, and the strains from Vietnam were also the cause of meningitis in adults. A unique DNA pattern of pulsotype D, classified into cluster XVI, was found for 3 strains with ST28 isolated from nonfatal cases in this study. Previous studies also reported 1 nonfatal case caused by the ST28 strain from Thailand and Japan (34,38).
Associations for bacteremic meningitis cases with nonfatal outcomes and probable meningitis cases with fatal outcomes contrasted strikingly in this study. Of 6 fatal cases of probable meningitis, 2 were caused by ST1, 2 by ST25, and 2 by ST104 strains. The extent to which the virulence of each ST strain contributed to these deaths remains uncertain. Another possible explanation may be a frequent involvement of critically ill patients, for whom lumbar puncture was not possible; these patients had probable meningitis and typical meningeal signs, acute disease onset, and positive blood culture only.
Because the clinical charts were retrospectively reviewed and the etiologic diagnosis of S. suis infection might not have been readily reported to the attending physicians during the hospitalization of the patients in this study, the extent of investigations of clinical manifestations, possible risk factors, and causes of death might have been limited. Because different physicians were involved in the assessment of different patients in this study, the possibility of misdiagnosis for clinical categories cannot be completely excluded even though meningeal signs and acute disease onset are clinical indicators of meningitis.
In conclusion, this study of the clinical features of 158 cases of S. suis seotype 2 infection in humans in Thailand showed that the disease occurs sporadically in adults and results in a mortality rate of ≈9.5%; the major clinical manifestations include meningitis and sepsis. MLST analyses of 165 isolates from humans indicated that the major STs were ST1 followed by ST104. Although both ST1 and ST104 strains cause sepsis, it is likely that only the ST1 strain causes meningitis. Further studies are needed to elucidate the pathogenesis of the human S. suis infections that are prevalent in Southeast Asian countries.
Mr Kerdsin is a molecular microbiologist at the National Institute of Health, Department of Medical Sciences, Ministry of Public Health, Thailand. His research interests include the molecular identification and epidemiology of bacterial pathogens, including S. suis.
Acknowledgments
We are grateful to the entire medical staff for their cooperation, especially N. Wongwan, H. Sudjit, T. Jaiwongsa, S. Amnajsirisuk, S. Chokngam, S. Boonyong, C. Noiyano, N. Singpolthan, C. Hiranyasuk, S. Worachuen, W. Joraka, S. Prasan, H. Phimartwisit, K. Katewong, C. Khumluang, B. Prawiset, U. Surin, C. Junethaworn, K. Kumisara, W. Noithachang, D. Amornthipayawong, S. Wangsai, T. Wongchai, T. Yeepu, and W. Pengreungrojanachai.
This work was supported research grants from the Department of Medical Sciences, Ministry of Public Health of Thailand, Grants-in Aid for Scientific Research (B: 21406027), and the program of Research Centers for Emerging and Reemerging Infectious Diseases launched by a project commissioned by the Ministry of Education, Science and Culture, and the Ministry of Health, Labor and Welfare of Japan.
References
- Lun ZR, Wang QP, Chen XG, Li AX, Zhu XQ. Streptococcus suis: an emerging zoonotic pathogen. Lancet Infect Dis. 2007;7:201–9. DOIPubMedGoogle Scholar
- Hill JE, Gottschalk M, Brousseau R, Harel J, Hemmingsen SM, Goh SH. Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotype 32 and 34 isolated from pigs, are Streptococcus orisratti. Vet Microbiol. 2005;107:63–9. DOIPubMedGoogle Scholar
- Ye C, Zhu X, Jing H, Du H, Seguera M, Zheng H, Streptococcus suis sequence type 7 outbreak, Sichuan, China. Emerg Infect Dis. 2006;12:1203–8.PubMedGoogle Scholar
- Wertheim HFL, Nghia HDT, Taylor W, Schultsz C. Streptococcus suis: an emerging human pathogen. Clin Infect Dis. 2009;48:617–25. DOIPubMedGoogle Scholar
- Fongcom A, Pruksakorn S, Mongkol R, Tharavichitkul P, Yoonim N. Streptococcus suis infection in northern Thailand. J Med Assoc Thai. 2001;84:1502–8.PubMedGoogle Scholar
- Wangkaew S, Chaiwarith R, Tharavichitkul P, Supparatpinyo K. Streptococcus suis infection: a series of 41 cases from Chiang Mai University Hospital. J Infect. 2006;52:455–60. DOIPubMedGoogle Scholar
- Rusmeechan S, Sribusara P. Streptococcus suis meningitis: the newest serious infectious diseases. J Med Assoc Thai. 2008;91:654–8.PubMedGoogle Scholar
- Wangsomboonsiri W, Luksananun T, Saksornchai S, Ketwong K, Sungkauparph S. Streptococcus suis infection and risk factors for mortality in northern Thailand. J Infect. 2008;57:392–6. DOIPubMedGoogle Scholar
- Khadthasrima N, Hannwong T, Thammawitjaya P, Pingsusean D, Akkanij B, Jaikhar A, Human Streptococcus suis outbreak in Phayao Province, Thailand, 2007. Outbreak, Surveillance, and Investigative Reports. 2009;1:4–7.
- Fongcom A, Prusakorn S, Netsirawan P, Pongprasert R, Onsibud P. Streptococcus suis infection: a prospective study in northern Thailand. Southeast Asian J Trop Med Public Health. 2009;40:511–7.PubMedGoogle Scholar
- Navacharoen N, Chabtharochavong V, Hanpasertpong C, Kangsanarak J, Lekagul S. Hearing and vestibular loss in Streptococcus suis infection from swine and traditional raw pork exposure in northern Thailand. J Laryngol Otol. 2009;123:857–62. DOIPubMedGoogle Scholar
- Kerdsin A, Oishi K, Sripakdee S, Boonkerd N, Polwichai P, Nakamura S, Clonal dissemination of Streptococcus suis serotype 14 in Thailand. J Med Microbiol. 2009;58:1508–13. DOIPubMedGoogle Scholar
- Mai NT, Hoa NT, Nga TV, Linh LD, Chau TT, Sinh DX, Streptococcus suis meningitis in adults in Vietnam. Clin Infect Dis. 2008;46:659–67. DOIPubMedGoogle Scholar
- Gottschalk M, Segura M. The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved question. Vet Microbiol. 2000;76:259–72. DOIPubMedGoogle Scholar
- Kim KS. Pathogenesis of bacterial meningitis: from bacteremia to neuron injury. Nat Rev Neurosci. 2003;4:376–85. DOIPubMedGoogle Scholar
- Vecht U, Wisselink HJ, Jellema ML, Smith HE. Identification of two proteins associated with virulence of Streptococcus suis type 2. Infect Immun. 1991;59:3156–62.PubMedGoogle Scholar
- Jacobs AA, Loeffen PLW, van den Berg AJG, Storm PK. Identification, purification, and characterization of a thiol-activated hemolysin (suilysin) of Streptococcus suis. Infect Immun. 1994;62:1742–8.PubMedGoogle Scholar
- Allen AG, Bolitho S, Lindsay H, Khan S, Bryant C, Norton P, Generation and characterization of a defined mutant of Streptococcus suis lacking suilysin. Infect Immun. 2001;69:2732–5. DOIPubMedGoogle Scholar
- Smith HE, Damman M, van der Velde J, Wagenaar JF, Wisselink HJ, Stockhofe-Zurwieden N, Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect Immun. 1999;67:1750–6.PubMedGoogle Scholar
- Baums CG, Kaim U, Fulde M, Ramachandran G, Goethe R, Valentin-Weigand P. Identification of a novel virulence determinant with serum opacification activity in Streptococcus suis. Infect Immun. 2006;74:6154–62. DOIPubMedGoogle Scholar
- Chen C, Tang J, Dong W, Wang C, Feng Y, Wang J, A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. [PMID: 1737520]. PLoS ONE. 2007;2:e315. DOIPubMedGoogle Scholar
- Li M, Wang C, Feng Y, Pan X, Cheng G, Wang J, SalK/SalR, a two-component signal transduction system, is essential for full virulence of highly invasive Streptococcus suis serotype 2. [PMID: 18461172]. PLoS ONE. 2008;3:e2080. DOIPubMedGoogle Scholar
- Marois C, Bougeard S, Gottschalk M, Kobisch M. Multiplex PCR assay for detection of Streptococcus suis species and serotypes 2 and 1/2 in tonsils of live and dead pigs. J Clin Microbiol. 2004;42:3169–75. DOIPubMedGoogle Scholar
- King SJ, Leigh JA, Heath PJ, Luque I, Tarradas C, Dowson CG, Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J Clin Microbiol. 2002;40:3671–80. DOIPubMedGoogle Scholar
- Rehm T, Baums CG, Strommenger B, Beyerbach M, Valentin-Weigand P, Goethe R. Amplified fragment length polymorphism of Streptococcus suis strains correlates with their profile of virulence-associated genes and clinical background. J Med Microbiol. 2007;56:102–9. DOIPubMedGoogle Scholar
- Feil EJ, Li BC, Anaensen DM, Hanage WP, Spatt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–30. DOIPubMedGoogle Scholar
- Silva LMG, Baums CG, Rehm T, Wisselink JR, Goethe R. P. Valentin-Weigand P. Virulence-associated gene profiling of Streptococcus suis isolates by PCR. Vet Microbiol. 2006;115:117–27. DOIPubMedGoogle Scholar
- Luey CKY, Chu YW, Cheung KM, Law CC, Chu MY, Cheung DT, Rapid pulsed-field gel electrophoresis protocol for subtyping of Streptococcus suis serotype 2. J Microbiol Methods. 2007;68:648–50. DOIPubMedGoogle Scholar
- McDaniel J, Pillai SD. Gel alignment and band scoring for DNA fingerprinting using Adobe Photoshop. Biotechniques. 2002;32:120–1, 123.PubMedGoogle Scholar
- Muchart DJ, Bhaganjee S. American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference definitions of the systemic inflammatory response syndrome and allied disorders in relation to critically injured patients. Crit Care Med. 1997;25:1765–95.PubMedGoogle Scholar
- Durack DT, Lukes AS, Bright DK. New criteria for diagnosis of infective endocarditis: utilization of specific echocardiographic findings. Am J Med. 1994;96:200–9. DOIPubMedGoogle Scholar
- Ferrer R, Artigas A, Levy MM, Blanco J, Gonzalez-Diaz G, Garnacho-Montero J, Improvement in process of care and outcome after a multicenter severe sepsis education program in Spain. JAMA. 2008;299:2294–303. DOIPubMedGoogle Scholar
- Takamatsu D, Wongsawan K, Osaki M, Nishino H, Ishii T, Thravichikul P, Streptococcus suis in humans, Thailand. Emerg Infect Dis. 2008;14:181–3. DOIPubMedGoogle Scholar
- Wertheim HF, Nguyen NH, Taylor W, Lien TT, Ngo HT, Nguyen TQ, Streptococcus suis, an important cause of adult bacterial meningitis in northern Vietnam. PLoS ONE. 2009;4:e5973. DOIPubMedGoogle Scholar
- Ma E, Chung PH, So T, Wong L, Choi KM, Cheung DT, Streptococcus suis infection in Hong Kong: an emerging infectious disease? Epidemiol Infect. 2008;136:1691–7. DOIPubMedGoogle Scholar
- Cheung P-Y, Lo KL, Cheung TT, Yeung WH, Leung PH, Yeung WH, Streptococcus suis in retail markets: how prevalent is it in raw pork? Int J Food Microbiol. 2008;127:316–20. DOIPubMedGoogle Scholar
- Chang B, Wada A, Ikebe T, Ohnishi M, Mita K, Endo M, Characteristics of Streptococcus suis isolated from patients in Japan. Jpn J Infect Dis. 2006;59:397–9.PubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 17, Number 5—May 2011
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Kazunori Oishi, Laboratory for Clinical Research on Infectious Diseases, International Research Center for Infectious Diseases, Research Institute for Microbial Diseases, Osaka University, 3-1 Yamadaoka, Suita 565-0871, Japan
Top