Volume 18, Number 1—January 2012
Dispatch
Spoligotyping of Mycobacterium africanum, Burkina Faso
Cite This Article
Citation for Media
Abstract
Using Ziehl-Neelsen–positive slides collected from tuberculosis diagnostic centers in Burkina Faso, we showed that 20% of 80 spoligotyping-positive DNA samples had a characteristic Mycobacterium africanum–specific genomic signature. This result suggests that M. africanum is still present in Burkina Faso at almost the same prevalence as 15–20 years ago.
Mycobacterium africanum remains a major pathogen in Africa (1). Recently, de Jong et al. estimated the prevalence of M. africanum to be 1.7% in Burkina Faso (1); their estimate was based on a 2007 study by Godreuil et al., who unexpectedly did not identify any M. africanum isolate within a collection of 120 M. tuberculosis complex clinical isolates in 2001 from 79 tuberculosis patients living in Ouagadougou and 41 living in Bobo Dioulasso, the 2 largest cities in the country (2). However, 2 patterns (isolates 94 and 90) can be recognized as M. africanum by their characteristic spoligotyping signature (deletion of spacers, 8, 9, and 39) and an association of mycobacterial interspersed repetitive unit (MIRU) 24 >2, MIRU31 >4, and MIRU40 <3 signature (2,3).
In the neighboring country of Ghana (which has 200 km of common borders with Burkina Faso), another study suggested that the population structure of M. tuberculosis complex comprises 1) 34% spoligo-international type (SIT) 61 (named the Cameroon clade, also present in Burkina Faso); 2) 30% M. africanum (including M. africanum West African 1 and West African 2); and 3) 36% principal genetic group 2 and 3 modern strains (e.g., T, U [unknown], Haarlem, X, LAM [Latino-American and Mediterranean]), with minor prevalence of other principal genetic groups, i.e., the East-African Indian, Beijing, and M. bovis clades (4,5). These observations—and their congruence to estimates by Ledru et al. in 1996 of an 18.4% prevalence of M. africanum strains isolated from the 300 patients in whom tuberculosis was newly diagnosed in Burkina Faso during 1992–1994 (5)—prompted us to reexamine the conclusions of Godreuil et al. on the M. africanum prevalence in Burkina Faso.
The study, which we conducted during March–September 2010, had 3 goals. First, we wanted to determine whether we could extract DNA and perform high-throughput spoligotyping on a Luminex 200 device (Luminex, Austin, TX, USA) on acid-fast bacillus–positive slides (6). Second, we wanted to reestimate the prevalence of M. africanum in Burkina Faso from a recent and random sample of slides. Third, we wanted to further analyze the relative proportion of M. africanum West African I and West African 2 strains in Burkina Faso because this country is part of central western Africa, where the 2 M. africanum West African 1 and 2 strains are present at various relative rates (2). We report on all the goals of this project, even though goal 3 remains to be confirmed because of the small sample size.
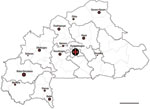
From within 14 geographically independent centers in Burkina Faso (Figure), we recruited a random sample of 186 Ziehl-Neelsen (ZN) slides that had been included in a national study on drug resistance, as approved by the ethical committee for health research in Burkina Faso (2007–031; June 28, 2009). Of 186 DNA samples extracted from as many ZN slides, 143 sputum samples had been scored 3+, 18 were scored 2+, 10 were scored 1+, 5 had 1–9 bacilli total (±), totaling 176 positive slides from as many sputum samples. In addition, test results were negative for 9 and unknown for 1.
In a preliminary trial of 9 independent 3+ positive slides, DNA extraction was attempted by 2 methods: an enzymatic method (7) and a classical thermic lysis in a Chelex suspension (InstaGene; Bio-Rad, Hercules, CA, USA) (8). In our study, only the Chelex method produced good results, i.e., enabled us to obtain DNA that was successfully PCR amplified and produced a full spoligotyping pattern (results not shown). The quantity of DNA extracted was superior for all tests by the enzymatic lysis (n = 3) as by the Chelex (n = 6), as estimated by spectrophotometry (NanoDrop ND-1000; LabTech, Ringmer, UK). Thus, DNA can be successfully extracted by the enzymatic method for many human or bacterial cells but not for M. tuberculosis complex because no spoligotype could be obtained. We therefore analyzed the 176 experimental slides by using the Chelex extraction procedure.
The origins of all ZN slides assessed in this study are shown in the Figure, and genotyping results are shown in the Table (full results in the Technical Appendix). As observed in a much larger set of samples from Ghana, the Cameroon family (SIT61) also prevails in Burkina Faso (18 [25%] isolates) (4). Whether the Cameroon strains from Burkina Faso are similar or identical to those from Ghana remains to be studied.
Our result confirms the observation by Godreuil et al. in 2007 on the prevalence of the SIT61/Cameroon strains in Burkina Faso (2). However, we detected 16 M. africanum spoligotypes (West African 1 and West African 2), i.e., a minimal M. africanum prevalence of 20%, close to 18.4% found by Ledru et al. in 1996 (5). Third, the T and Haarlem strains represented 16 (22%) and 10 (14%), respectively, of the patterns; other genotypes were rare (5 CAS [Central Asian], 4 X, 1 M. bovis, 1 Beijing, 1 LAM). Finally, the relative prevalence of M. africanum West African 1 from M. africanum West African 2 could first be assessed by the spoligotyping signature (2 vs. 14; Technical Appendix). Specific single nucleotide polymorphism detection could constitute another classification tool for M. africanum sublineages (10,11). Unfortunately, detection of katG203 single nucleotide polymorphism failed on the slide-extracted DNAs (results not shown), and our study is limited by a suboptimal yield in positive spoligotyping results (80 [43%] of 186), an issue that should be improved.
The results of our study diverge on the M. africanum prevalence in Burkina Faso from results from Godreuil et al. (2) (1.9% vs. 20%). These authors were intrigued to not identify more M. africanum isolates and suggested that their finding might reflect “a decrease in M. africanum prevalence in these countries,” referring to a similar decrease in Cameroon during 1971–2003 (12,13). We believe that in the study by Godreuil et al., an unintentional bias was introduced against M. africanum, given the difficulty of isolating this genotypic variant in routine practice in mycobacteriologic laboratories. Differences in M. africanum prevalence in culture-based and sputum-based studies might reflect the difficulties of growing and isolating M. africanum in some national TB reference laboratories in western Africa. M. africanum, which is closely related to M. bovis, has peculiar growing requirements that are not always satisfied. Supplementation of Löwenstein-Jensen medium with pyruvate is mandatory and not standardized (from 0.1% to 0.4%).
The pyruvate requirements of some members of the M. tuberculosis complex were recently shown to be caused by a mutation creating an inactive pyruvate kinase (14). This specific mutation of M. africanum has major implications for its metabolism and growth.
Implementation of adequate culture and molecular identification facilities in Burkina Faso are needed. A potential solution to avoiding the bias from culture and from DNA extraction from slides could be to extract DNA directly from sputum, e.g., by storing surplus sputum prospectively in 70% ethanol. Additional work also is needed to improve analytical methods for ZN slides to refine description of M. tuberculosis genetic diversity and eventually to provide predictive genetic drug susceptibility testing. Introduction of newer and faster TB diagnostic methods are urgently needed in this area of western Africa.
Dr Gomgnimbou is a public health scientist of the Centre Muraz in Bobo Dioulasso, Burkina Faso, and is following a doctoral program of the École Doctorale Gènes Génomes Cellules of the University Paris-Sud 11 working in the Infection Genetics Emerging Pathogens Evolution Team at the Institute of Genetics and Microbiology in Orsay, France. His research interests focus on new diagnostics and genotyping methods applicable to tuberculosis control.
Acknowledgments
We are grateful to M. Daouda for producing the Burkina Faso map and to M. François Topin for technical support.
The Global Fund and West Africa Network of Excellence for Tuberculosis, HIV/AIDS, and Malaria (European and Developing Countries Clinical Trials Partnership) helped fund this study. This work was made possible through the financial support of the French Embassy in Burkina Faso (Services d’Action Culturelle et de Coopération) through a Masters-level study grant to M.K.G.
References
- de Jong BC, Antonio M, Gagneux S. Mycobacterium africanum–review of an important cause of human tuberculosis in west Africa. PLoS Negl Trop Dis. 2010;4:e744. DOIPubMedGoogle Scholar
- Godreuil S, Torrea G, Terru D, Chevenet F, Diagbouga S, Supply P, First molecular epidemiology study of Mycobacterium tuberculosis in Burkina Faso. J Clin Microbiol. 2007;45:921–7. DOIPubMedGoogle Scholar
- Viana-Niero C, Gutierrez C, Sola C, Filliol I, Boulahbal F, Vincent V, Genetic diversity of Mycobacterium africanum clinical isolates based on IS6110-restriction fragment length polymorphism analysis, spoligotyping, and variable number of tandem DNA repeats. J Clin Microbiol. 2001;39:57–65. DOIPubMedGoogle Scholar
- Intemann CD, Thye T, Niemann S, Browne EN, Amanua Chinbuah M, Enimil A, Autophagy gene variant IRGM −261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog. 2009;5:e1000577. Epub 2009 Sep 11. DOIPubMedGoogle Scholar
- Ledru S, Cauchoix B, Yameogo M, Zoubga A, Lamande-Chiron J, Portaels F, Impact of short-course therapy on tuberculosis drug resistance in south-west Burkina Faso. Tuber Lung Dis. 1996;77:429–36. DOIPubMedGoogle Scholar
- Zhang J, Abadia E, Refregier G, Tafaj S, Boschiroli ML, Guillard B, Mycobacterium tuberculosis complex CRISPR genotyping: improving efficiency, throughput and discriminative power of “spoligotyping” with new spacers and a microbead-based hybridization assay. J Med Microbiol. 2010;59:285–94. DOIPubMedGoogle Scholar
- Kamble RR, Shinde VS, Madhale SP, Kamble AA, Ravikumar BP, Jadhav RS. Extraction and detection of Mycobacterium leprae DNA from ZNCF-stained skin smear slides for better identification of negative skin smears. Indian J Med Microbiol. 2010;28:57–9. DOIPubMedGoogle Scholar
- van der Zanden AG, Hoentjen AH, Heilmann FG, Weltevreden EF, Schouls LM, van Embden JD. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis complex in paraffin wax embedded tissues and in stained microscopic preparations. Mol Pathol. 1998;51:209–14. DOIPubMedGoogle Scholar
- Brudey K, Driscoll J, Rigouts L, Prodinger WM, Gori A, Al-Hajoj SAM, Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, Population Genetics, and Epidemiology. BMC Microbiol. 2006;6:23. DOIPubMedGoogle Scholar
- Frothingham R, Strickland PL, Bretzel G, Ramaswamy S, Musser JM, Williams DL. Phenotypic and genotypic characterization of Mycobacterium africanum isolates from west Africa. J Clin Microbiol. 1999;37:1921–6.PubMedGoogle Scholar
- Vasconcellos SE, Huard RC, Niemann S, Kremer K, Santos AR, Suffys PN, Distinct genotypic profiles of the two major clades of Mycobacterium africanum. BMC Infect Dis. 2010;10:80. DOIPubMedGoogle Scholar
- Huet M, Rist N, Boube G, Potier D. Etude bactériologique de la tuberculose au Cameroon. Rev Tuberc Pneumol (Paris). 1971;35:413–26.PubMedGoogle Scholar
- Niobe-Eyangoh SN, Kuaban C, Sorlin P, Cunin P, Thonnon J, Sola C, Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J Clin Microbiol. 2003;41:2547–53. DOIPubMedGoogle Scholar
- Keating LA, Wheeler PR, Mansoor H, Inwald JK, Dale J, Hewinson RG, The pyruvate requirement of some members of the Mycobacterium tuberculosis complex is due to an inactive pyruvate kinase: implications for in vivo growth. Mol Microbiol. 2005;56:163–74. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleTable of Contents – Volume 18, Number 1—January 2012
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Christophe Sola, Institut de Génétique et Microbiologie– Infection Genetics Emerging Pathogens Evolution Team, Campus d’Orsay, Bldg 400, Rm 205, Orsay Ile de France F-91405
Top