Volume 18, Number 10—October 2012
CME ACTIVITY - Research
Methicillin-Resistant Staphylococcus aureus Sequence Type 239-III, Ohio, USA, 2007–20091
Cite This Article
Citation for Media
Introduction

MEDSCAPE CME
Medscape, LLC is pleased to provide online continuing medical education (CME) for this journal article, allowing clinicians the opportunity to earn CME credit.
This activity has been planned and implemented in accordance with the Essential Areas and policies of the Accreditation Council for Continuing Medical Education through the joint sponsorship of Medscape, LLC and Emerging Infectious Diseases. Medscape, LLC is accredited by the ACCME to provide continuing medical education for physicians.
Medscape, LLC designates this Journal-based CME activity for a maximum of 1 AMA PRA Category 1 Credit(s)TM. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
All other clinicians completing this activity will be issued a certificate of participation. To participate in this journal CME activity: (1) review the learning objectives and author disclosures; (2) study the education content; (3) take the post-test with a 70% minimum passing score and complete the evaluation at www.medscape.org/journal/eid; (4) view/print certificate.
Release date: September 20, 2012; Expiration date: September 20, 2013
Learning Objectives
Upon completion of this activity, participants will be able to:
• Distinguish the most common strains of methicillin-resistant Staphylococcus aureus (MRSA) in the United States
• Assess the clinical characteristics of infection with MRSA-ST239-III
• Analyze the treatment and prognosis of MRSA-ST239-III infection
• Evaluate molecular characteristics of MRSA-ST239-III.
CME Editor
Thomas J. Gryczan, MS, Technical Writer/Editor, Emerging Infectious Diseases. Disclosure: Thomas J. Gryczan, MS, has disclosed no relevant financial relationships.
CME Author
Charles P. Vega, MD, Health Sciences Clinical Professor; Residency Director, Department of Family Medicine, University of California, Irvine. Disclosure: Charles P. Vega, MD, has disclosed no relevant financial relationships.
Authors
Disclosures: Shu-Hua Wang, MD, MPH; Yosef Khan, MBBS, PhD; Jose R. Mediavilla, MBS, MPH; Liangfen Zhang, MD, PhD; Liang Chen, PhD; Armando Hoet, PhD; Tammy Bannerman; D. Ashley Robinson, PhD; Barry N. Kreiswirth, PhD; and Kurt B. Stevenson, MD, MPH, have disclosed no relevant financial relationships. Lisa Hines, RN, CIC, has disclosed the following relevant financial relationships: owns stock, stock options, or bonds from Kimberly-Clark Corp., General Electric Co., Medtronic Inc., Stryker Corp., TEVA Pharmaceutical Industries. Preeti Pancholi, PhD, has disclosed the following relevant financial relationships: served as an advisor or consultant for Abbott; served as a speaker or a member of a speakers bureau for Abbott, Nanosphere; received grants for clinical research from Cepheid, Abbott, Quidel, Qiagen, Nanosphere.
Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is a human pathogen that has diverse molecular heterogeneity. Most MRSA strains in the United States are pulsed-field gel electrophoresis USA100 sequence type (ST) 5 and USA300 ST8. Infections with MRSA ST239-III are common and found during health care–associated outbreaks. However, this strain has been rarely reported in the United States. As part of a study supported by the Prevention Epicenter Program of the Centers for Disease Control and Prevention (Atlanta, GA, USA), which evaluated transmission of MRSA among hospitals in Ohio, molecular typing identified 78 (6%) of 1,286 patients with MRSA ST239-III infections. Ninety-five percent (74/78) of these infections were health care associated, and 65% (51/78) of patients had histories of invasive device use. The crude case-fatality rate was 22% (17/78). Identification of these strains, which belong to a virulent clonal group, emphasizes the need for molecular surveillance.
Staphylococcus aureus is a major human pathogen that possesses multiple toxins and virulence mechanisms (1). Antimicrobial drug resistance in S. aureus has added to the complexity of treating serious infections caused by this bacteria, and methicillin-resistant S. aureus (MRSA) appears to have greater virulence than methicillin-susceptible strains (2,3). Most MRSA strains in the United States are pulsed-field gel electrophoresis (PFGE) types USA100 and USA300, corresponding to multilocus sequence typing (MLST) ST5 and ST8, respectively (4). MRSA belonging to MLST ST239 and harboring staphylococcal cassette chromosome mec (SCCmec) type III (MRSA ST239-III) are associated with infections in health care settings, outbreaks, increased resistance to antimicrobial drugs, and capacity for invasive disease (5–7).
MRSA ST239-III has a history of successful dissemination in many regions, leading to a diverse array of regionally prevalent clones. These clones include the Brazilian; British Epidemic 1, 4, 7, 9, and 11; Canadian Epidemic 3/Punjab; Czech; Eastern Australian 2 and 3; Georgian; Hungarian; Lublin; Nanjing/Taipei (ST241); Portuguese; and Vienna clones (8,9). Although it is common worldwide, MRSA ST239-III has not played any predominant role in the United States; infections with MRSA ST239-III have been rarely reported in the United States since the 1990s (9–13). Recently, only 2 reports of this strain in the United States involving sporadic nasal colonization and bloodstream infections have been published (13,14).
In this study, we describe clinical epidemiologic characteristics and molecular analysis of clinical infections with MRSA ST239-III in the midwestern United States. Identification of a strain from such a virulent clonal group in the United States with wide dissemination in other parts of the world represents a potential public health concern.
Sampling Method
As part of Centers for Disease Control and Prevention (CDC) (Atlanta, GA, USA)–sponsored Prevention Epicenter Program study evaluating the transmission of MRSA between hospitals in Ohio, USA, molecular analysis was performed on a group of clinical MRSA isolates collected from The Ohio State Health Network (OSHN). The OSHN consists of The Ohio State University (OSU) Wexner Medical Center (WMC), which is a tertiary care medical center, and 7 smaller community hospitals located 30–120 miles from OSU. All MRSA specimens from community hospitals and selected OSU MRSA blood isolates and isolates from patients residing in the catchment areas of the outreach hospitals were prospectively collected during March 2009–February 2010 for genotyping by using a repetitive element PCR (rep-PCR). Among archived MRSA isolates from OSUWMC from January 2007 through February 2009, only a selection of isolates was chosen for genotyping (not a randomized sampling). The total number of isolates and the total number of ST239 from each time period was collected.
Data Collection
We performed medical record reviews for 1,286 patients. Patient demographic characteristics (presence of health care–related risk factors during the preceding 12 months, presence of an invasive device during the previous 7 days, and concurrent conditions) were collected. Patient-level data, including addresses for geocoding, were entered into a secure database within the OSUWMC Information Warehouse.
Classification of MRSA Infections
All MRSA cases were classified into 3 categories on the basis of accepted epidemiologic definitions (11). The first category was health care–associated, defined as a culture obtained >48 hours after admission. The second category was health care–associated community onset, defined as a culture obtained <48 hours after admission with identified health care–associated risk factors. The third category was community-associated, defined as a culture obtained <48 hours after admission without health care–associated risk factors. Health care–associated risk factors comprised presence of an invasive device, history of MRSA infection or colonization, surgery, hospitalization, dialysis, or residence in a long-term care facility in the 12 months preceding the culture.
Outcomes for MRSA infection were categorized as cure (complete resolution after antimicrobial drug treatment); failure (persistence of infection and change in antimicrobial drug regimen); relapse (resolution of infection after complete treatment with subsequent development of new symptoms); recurrent (redevelopment of MRSA at same or other site >2 weeks after completion of treatment for initial MRSA infection); indeterminate (unknown outcome); and death (death <30 days after diagnosis of MRSA infection because of any cause or during the same hospitalization). Destination after hospital discharge, such as home or skilled nursing facility, was also noted.
Drug Susceptibility Testing
The respective OSHN Clinical Microbiology Laboratories initially identified all MRSA isolates by using standard microbiological methods. Antimicrobial drug susceptibility testing was performed at each institution, and results were interpreted according to Clinical and Laboratory Standards Institute break point guidelines (15). At OSUWMC, antimicrobial drug susceptibility testing was performed by using the automated Micro-Scan method (Siemens Diagnostics, Sacramento, CA, USA), and only constitutive clindamycin testing was performed. Linezolid MIC >4 mg/L were confirmed by using the Etest method (bioMérieux, Marcy l’Etoile, France).
Genotyping
The MRSA ST239 III isolates were genotyped initially by using rep-PCR, followed by PFGE, staphylococcal protein A sequencing (spa typing), SCCmec typing, and mec-associated direct repeat unit (dru) typing. Selected isolates were also characterized by MLST and single-nucleotide polymorphism (SNP) typing. Detection of genes encoding Panton–Valentine leukocidin (PVL), toxic shock syndrome toxin (TSST), arginine catabolic mobile element (ACME), and high-level mupirocin resistance (mupA) was also performed. Brief descriptions of each testing method are outlined below.
rep-PCR
The DiversiLab System (bioMérieux, Durham, NC, USA) was used for rep-PCR analysis according to described methods (16). Isolates belonging to designated rep-PCR clusters shared >95% similarity. In addition, comparison of matching patterns in the DiversiLab System library was initially used to infer the PFGE and SCCmec types, which were later validated by using appropriate testing methods. The numeric classification system used for rep-PCR analysis is unique to OSUWMC.
PFGE
The PulseNet protocol for molecular subtyping of S. aureus was followed. Salmonella enterica serotype Braenderup DNA was digested with XbaI (Roche, Indianapolis, IN, USA) and used as the normalization standard for gel analysis. S. aureus chromosomal DNA was digested with SmaI (Roche). Fragments were separated in a clamped homogenous electric field mapper unit (Bio-Rad Laboratories, Hercules, CA, USA). Fingerprint images were analyzed by using Bionumerics software version 4.61 (Applied Maths NV, Sint-Martens-Latem, Belgium). The traditional classification of PFGE subtypes was not used because we were not analyzing for an outbreak (17). Thus, interpretations of possibly or probably related PFGE subtypes between strains obtained by PFGE band patterns were not made. Each PFGE band difference was classified as a unique PFGE pattern.
spa Typing
spa typing (18) was performed on all MRSA isolates by using eGenomics software (www.egenomics.com) as described (19); Ridom spa types were subsequently assigned by using the SpaServer website (www.spaserver.ridom.de).
SCCmec Typing
SCCmec typing was performed on all MRSA isolates by using a described multiplex real-time PCR (20). This PCR is specific for 2 essential gene complexes (ccr and mec) found in all SCCmec elements.
dru Typing
MRSA isolates were also characterized by sequencing the hypervariable dru repeat region within the SCCmec element (21) and using DruID software (9). New dru types were submitted to www.dru-typing.org.
MLST and PVL, TSST, ACME, and Mupirocin Resistance
MLST was performed on representative isolates as described (22) by using the MLST database (http://saureus.mlst.net). PCR-based detection of PVL (23), TSST (24), ACME (25), and mupA (26) was performed on all isolates as described.
SNP Typing
A panel of 43 SNPs for describing the global population structure of MRSA ST239-III (9) was used to identify the haplotypes of 22 isolates. These SNPs were typed by using Golden-Gate Genotyping Assay (Illumina, San Diego, CA, USA) and conventional Sanger sequencing.
Statistical Analysis
All patient demographic, clinical, and molecular typing data were aggregated in tabular format, and descriptive statistics were generated by using SAS version 9.2 (SAS Institute Inc., Cary NC, USA), for demographic and risk factor history. A significant difference between ST239 and other strains (US300, US100, and all other strains) was examined by using χ2 tests for categorical variables and t-tests for continuous variables. An α level of 0.5 was used.
Human Subjects Protection
We obtained approval for this study from the OSU Office of Responsible Research Practices’ Biomedical Institutional Review Board. The OSU Information Warehouse has established honest broker status with the OSU Institutional Review Board, enabling storage of fully identifiable data and presentation of patient data to investigators in a coded format that maintains patient confidentiality.
Patient Characteristics
Of 1,286 clinical MRSA isolates, 78 (6%) were identified as MRSA ST239-III; 71 (91%) were obtained from OSU and 7 (9%) from community hospitals. Seven (2%) of 397 isolates were obtained from outreach community hospitals, 37 (6%) of 613 from OSUWMC, and 34 (12%) of 276 from OSUWMC archives.
These strains were first recognized by rep-PCR as possible SCCmecA type III isolates from the DiversiLab System library with imputed Brazilian PFGE types. Additional molecular typing identified MRSA ST239-III in a clade different from that containing the Brazilian strains.
Demographics and clinical characteristics of 78 patients infected with MRSA ST239-III are shown in the Table. Among patients with MRSA ST239-III infections, 46 (59%) were male, 65 (83%) were white, 31 (40%) disabled, and 29 (37%) were retired. Seventy-four (95%) had health care–associated isolates and 51 (65%) had histories of use of invasive devices within the previous 7 days. Distribution of specimen types include 37 (47%) bloodstream infections (BSIs), 19 (24%) lower respiratory tract infections (LRTIs), 11 (14%) skin and soft tissue infections, and 11 (14%) other infections. Seventeen (22%) patients died, and 18 (23%) patients had treatment failures, recurring infections, or relapses. Only 32% of the patients were discharged home.
Comparison of clinical characteristics of MRSA ST239-III with those of PFGE types USA100 and USA300 and other non–ST239 infections are shown in the Table. Most ST239 isolates were health care–associated MRSA and had characteristics and concurrent conditions similar to those associated with USA100. MRSA ST239-III did not have virulent determinants often associated with health care–associated or community-associated MRSA strains, such as PVL, TSST, ACME, or mupA.
Characteristics of MRSA ST239-III Isolates
Resistance was observed to clindamycin (76/76, 100%), moxifloxacin (47/47, 100%), gentamicin (74/77, 96%), tetracycline (70/74, 95%), and trimethoprim–sulfamethoxazole (62/77, 81%). All MRSA ST239-III isolates were susceptible to vancomycin. For daptomycin, 64 (98%) of 65 isolates were susceptible, and 1 blood isolate with an MIC of 2 mg/L was classified as uninterpretable. For linezolid, 69 (97%) of 71 isolates were susceptible and 2 blood isolates with MICs >4 mg/L were classified as uninterpretable.
Molecular Typing of MRSA ST239-III Isolates
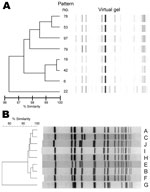
Eight rep-PCR patterns (6, 19, 22, 42, 53, 78, 79, and 97) were detected among MRSA ST239-III isolates, and 39 (50%) isolates had rep-PCR pattern 6. Similarly, isolates were distributed among 9 PFGE patterns (A, B, C, D, E, F, G, H, and I), and 58 (74%) had PFGE pattern A. The rep-PCR and PFGE patterns are shown in Figure 1. PFGE pattern F is <80% similar with other PFGE subtypes in the dendrogram and was identified as an unknown PFGE type by the CDC database. However, molecular testing confirmed it to be MRSA ST239-III: MLST 239, SCCmec type III, spa type 3(t037), dru type dt15b, and SNP haplotype H9.
Four spa types, including eGenomics spa types 3 (Ridom t037), 314 (t363), 121 (t421), and 1256 (t631) were identified, and 74 (95%) isolates were classified as spa type 3 (t037). In contrast, isolates were grouped into 13 dru types (dt2c, dt6n, dt9g, dt12i, dt13k, dt14 g, dt15b, dt15h, dt15i, dt15j, dt15k, dt16a, and dt23a), and 60 (77%) isolates were classified as dt15b. MLST typing of selected representative isolates confirmed 13 to be MRSA ST239-III (2-3-1-1-4-4-3), 3 as ST1801 (2-3-1-1-4-19-3), and 1 as ST2017 (2-3-1-1-4-19-227). ST1801 is a single-locus variant of MRSA ST239-III, and ST2017 is a novel single-locus variant of ST1801 and a double-locus variant of MRSA ST239. All were SCCmec type III, and the ccrC locus was not detected. The genes that encode PVL, TSST, ACME, or high-level mupirocin resistance were not found in any isolates.
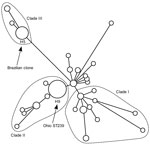
A total of 33 unique genotypic combinations from 78 patients infected with MRSA ST239-III were represented among isolates with complete SCCmec, rep-PCR, PFGE, spa, and dru data. This information and year of isolation are shown in Figure 2. The genotype cluster sizes ranged from 2 to 18 isolates; 23 of the isolates were unique. In contrast, SNP typing of 22/78 isolates with a panel of 43 SNPs showed them to be indistinguishable from each other. All isolates tested belonged to haplotype 9 (H9) within MRSA ST239-III clade II (Figure 3). This clade was composed of isolates from many continents, including many from sources in Asia (9).
MRSA ST239-III has demonstrated epidemic potential worldwide, and identification of this strain in the United States might represent a major public health concern. Although the precise time frame of emergence of this strain in Ohio is unknown, it appears to have been present before the study because it was identified in archived specimens dating back to 2007. The origin of the MRSA ST239-III clonal group has been dated to the mid-20th century in 2 studies (9,27). Thus, the Ohio strain might have been introduced into the region anytime during the past 40 years. Because of incomplete sampling during the study, the transmission dynamics of Ohio MRSA ST239 III are uncertain. Whether the prevalence would have been >6% if complete sampling was conducted is unknown. Furthermore, we found no epidemiologic evidence of clustering of cases consistent with an outbreak of infection with this strain. The large number of isolates at OSUWMC merely reflects its role as a large referral center.
Clinically, MRSA ST239-III is primarily health care associated, causes major outbreaks, shows increased resistance to antimicrobial drugs, and demonstrates a capacity for causing invasive disease (5–7,28,29). Comparison of characteristics of patients infected with MRSA ST239-III with those infected with PFGE USA300, PFGE USA100, and other strains showed that MRSA ST239-III is similar to USA100 in terms of health care–associated risk factors and outcomes (Table).
Concern for the invasive potential of MRSA ST239-III was illustrated during a 2-year intensive care unit (ICU) outbreak in London in which patients infected with MRSA ST239-III were more likely than patients with non–ST239 strains to show development of bacteremia (47% vs 13%; p<0.001) and have culture-positive vascular access device infections (59% vs 26%; p<0.001) (29). In our study, 19% (14/74) of patients infected with MRSA ST239-III strains were admitted to ICU, compared with 11% (96/850) of patients infected with non–ST239 strains (Table). Among MRSA ST239-III ICU admissions, 50% (7/14) were BSIs and 50% (7/14) were LRTIs. The crude mortality rate for persons with BSIs and LRTIs was 22% (8/37) and 47% (9/19) respectively. In our study population, MRSA ST239-III had a higher case-fatality rate than did USA300 (4%) and USA100 (17%) (Table).
Increased illness and death for pulmonary disease might be caused by an enhanced ability of MRSA ST239-III strains to produce biofilms and adhere to airway epithelial cells, providing a biologically plausible explanation for its potential to cause pneumonia and central line–associated BSIs (28,29). The association of MRSA ST239-III isolates with pulmonary infections has also been reported in South Korea (30). Moreover, in a multicenter study involving 21 hospitals in Beijing, where MRSA ST239-III is especially prevalent, 61% of MRSA LRTI cases were attributed to MRSA ST239-III (31). The London outbreak strain of MRSA ST239-III, and most examined MRSA ST239-III strains from the People’s Republic of China, belong to the same clade (clade II) as the Ohio strain of MRSA ST239-III, demonstrating the potential for this lineage to disseminate and cause disease (9,27,32). Because of incomplete collection of all MRSA strains causing LRTIs, whether MRSA ST239-III is the prevalent strain in MRSA pneumonias is unknown. A total of 17% (219/1,286) of the study isolates were pulmonary infections and 9% (19/219) of the pneumonias were caused by MRSA ST239-III.
PFGE is the standard for MRSA outbreak investigations. However, we chose a rapid PCR–based method that could serve as a potential screening surveillance tool to identify outbreaks of a particular strain. Although many institutions might be using PFGE for outbreak investigations to identify clonal clusters, methods for PFGE subtype classification are often not standardized between laboratories (33,34). To maximize uniformity and enable comparisons of PFGE classifications between institutions, the standardized CDC protocols for performing and analyzing PFGE results were used. The initial CDC PFGE analysis classified our isolates as the Brazilian clone. However, SNP typing unambiguously placed the Ohio isolates within a different clade than the Brazilian clone (Figure 3) (9). Because PFGE typing cannot define specific lineages for MRSA ST239-III, CDC has changed the reporting of MRSA ST239-III strains from Brazilian subtype to ST239. Laboratories will need to perform additional testing to identify specific clades or clonal groups. Classification of the 78 MRSA ST239-III isolates by the traditional PFGE outbreak method would identify only 2 PFGE subtypes instead of the 8 observed patterns. PFGE subtype 1 would consist of isolates A–E, G, and H, and subtype 2 would consist of isolate F (Figure 1, panel B).
Classification issues also exist with respect to the DiversiLab system. Five of the 8 initial rep-PCR patterns matched isolates in the DiversiLab library, which likewise categorized them as the Brazilian clone. In contrast, 3 other rep-PCR patterns did not match any isolates in the DiversiLab library. Additional molecular tests were performed to identify these isolates as MRSA ST239-III.
Because of the novel identification of MRSA ST239-III strains, we performed a battery of molecular tests to better classify the strains. Although the same dru types might be found in unrelated MRSA lineages and different staphylococcal species (32), variation at the dru locus within the SCCmec III element is consistent with MRSA ST239-III phylogeny (9). Because use of this typing method is relatively new, further study of its phylogenetic utility is needed (21). The feasibility of population-based genome-wide SNP datasets has been demonstrated for MRSA ST239-III by Harris et al. (27). Cluster groupings from these PFGE, rep-PCR, and dru methods were not identical, suggesting that the molecular tests are not completely interchangeable. Of the various molecular methods used to index genomic variation in MRSA, dru typing and rep-PCR appeared to be more discriminatory.
The publication of only 2 recent reports of MRSA ST239-III in the United States (13,14) might have resulted from inadequate national surveillance or low transmissibility of the strain. Institutions might not be able to perform genotyping on all their isolates because of lack of appropriate laboratory facilities or resources. An alternative method of surveillance for MRSA ST239-III might be evaluation of the drug susceptibility pattern. Most of the MRSA ST239-III strains in our study were resistant to clindamycin, tetracycline, and gentamicin. In a subset analysis, the phenotypic patterns of antimicrobial drug susceptibility and MRSA genotype prediction were determined for 798 MRSA isolates from the OSHN dataset. Ninety-four percent (63/67) of the MRSA ST239-III isolates were resistant to all 3 drugs (clindamycin, tetracycline, and gentamicin) (35). Use of phenotypic drug susceptibility patterns might alert infection control practitioners that they are dealing with a MRSA ST239-III clone. Additional molecular testing, such as spa typing in conjunction with SCCmec typing, can then be performed to confirm the strain type.
We have genotyped and geocoded 1,286 MRSA isolates in the CDC Prevention Epicenter Study by using rep-PCR. The lack of a clear association between MRSA ST239-III molecular genotype patterns could be caused by the retrospective nature of this study, lack of complete population sampling, and the inability to gather social network history. The rep-PCR and PFGE methods might be of limited use in evaluating geographic clustering at the scale studied here unless specific lineages of the reference strains are included in the respective databases. Because of the clonality exhibited by MRSA ST239-III, even a relatively small panel of 43 SNPs is able to identify the same major phylogenetic lineages that are identified by genome-wide SNPs. A more detailed genome-wide SNP analysis might be required to resolve geographic clustering. Additional social networking for determining spatial, temporal, and geographic relationships of patients at the medical institutions studied is under way to identify potential nosocomial interhospital and intrahospital transmission.
Recent identification of a strain of MRSA ST239-III in the midwestern United States is a major public health concern. Globally, this strain has demonstrated increased virulence and widespread dissemination. Because of an inadequate national surveillance system, temporal emergence and dissemination of the strain in the United States is uncertain. The MRSA ST239-III strains in Ohio have molecular heterogeneity and geographic diversity. The SNP-based data suggest that the strains also show clonal diversity. Continued surveillance is warranted because this MRSA ST239-III strain, similar to related strains worldwide, exhibits increased antimicrobial drug resistance, capacity for causing invasive disease, potential for causing outbreaks, and resulting in illnesses and deaths. MRSA ST239-III strains might have the potential to become established locally and disseminate among health care institutions as reported in other regions. Our study underscores the value of molecular surveillance, including traditional fingerprinting methods and newer sequence-based typing methods, as a critical component in understanding the evolving epidemiology of MRSA.
Dr Wang is an assistant professor of medicine at The Ohio State University in Columbus, Ohio. Her research interests are molecular genotyping, geocoding, and social network analysis for evaluation of transmission of MRSA and Mycobacterium tuberculosis.
Acknowledgments
We thank the OSU Epicenter Program Team, OSHN, for assistance with data collection; CDC for assistance with initial SCCmec typing; Gregory Fosheim for assistance with PFGE and SCCmec typing; Jennifer Santangelo, David Newman, and Kelly Kent for assistance with the Web entry portal and data management; Ruchi Tiwari for rep-PCR typing; Eric Brandt for PFGE typing; Melissa Abley and Brandon Palinski for MLST typing; and Rachael Tumin for phenotypic analysis.
S.-H.W. was supported by OSU Centers for Clinical Translational Science and the Marie Davis Bremer Award and by Award KL2 RR02574 from the National Centers for Research Resources. D.A.R. was supported by a grant from the American Heart Association and National Institutes of Health grant GM080602. S.-H.W., Y.K, L.H. P.P., and K.B.S. were supported by CDC Prevention Epicenter program grant (U01 CI000328).
References
- Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis. 2003;36:53–9. DOIPubMedGoogle Scholar
- Melzer M, Eykyn SJ, Gransden WR, Chinn S. Is methicillin-resistant Staphylococcus aureus more virulent than methicillin-susceptible S. aureus? A comparative cohort study of British patients with nosocomial infection and bacteremia. Clin Infect Dis. 2003;37:1453–60. DOIPubMedGoogle Scholar
- McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41:5113–20. DOIPubMedGoogle Scholar
- Feil EJ, Nickerson EK, Chantratita N, Wuthiekanum V, Srisomang P, Cousind R, Rapid detection of the pandemic methicillin-resistant Staphylococcus aureus clone ST 239, a dominant strain in Asian hospitals. J Clin Microbiol. 2008;46:1520–2. DOIPubMedGoogle Scholar
- Holden MT, Lindsay JA, Corton C, Quail MA, Cockfield JD, Pathak S, Genome sequence of a recently emerged, highly transmissible, multi-antibiotic- and antiseptic-resistant variant of methicillin-resistant Staphylococcus aureus, sequence type 239 (TW). J Bacteriol. 2010;192:888–92. DOIPubMedGoogle Scholar
- Aires de Sousa M, Conceição T, Simas C, de Lencastre H. Comparison of genetic backgrounds of methicillin-resistant and -susceptible Staphylococcus aureus isolates from Portuguese hospitals and the community. J Clin Microbiol. 2005;43:5150–7. DOIPubMedGoogle Scholar
- Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE. 2011;6:e17936. DOIPubMedGoogle Scholar
- Smyth DS, McDougal LK, Gran FW, Manoharan A, Enright MC, Song JH, Population structure of a hybrid clonal group of methicillin-resistant Staphylococcus aureus, ST239-MRSA-III. PLoS ONE. 2010;5:e8582. DOIPubMedGoogle Scholar
- de Lencastre H, Severina EP, Roberts RB, Kreiswirth BN, Tomasz A. Testing the efficacy of a molecular surveillance network: methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecium (VREF) genotypes in six hospitals in the metropolitan New York City area. The BARG Initiative Pilot Study Group. Bacterial Antibiotic Resistance Group. Microb Drug Resist. 1996;2:343–51. DOIPubMedGoogle Scholar
- Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–71. DOIPubMedGoogle Scholar
- Chung M, Dickinson G, de Lencastre H, Tomasz A. International clones of methicillin-resistant Staphylococcus aureus in two hospitals in Miami, Florida. J Clin Microbiol. 2004;42:542–7. DOIPubMedGoogle Scholar
- Tenover FC, Tickler IA, Goering RV, Kreiswirth BN, Mediavilla JR, Persing DH. Characterization of nasal and blood culture isolates of methicillin-resistant Staphylococcus aureus from patients in United States hospitals. Antimicrob Agents Chemother. 2012;56:1324–30. DOIPubMedGoogle Scholar
- Mermel LA, Eells SJ, Acharya MK, Cartony JM, Dacus D, Fadem S, Quantitative analysis and molecular fingerprinting of methicillin-resistant Staphylococcus aureus nasal colonization in different patient populations: a prospective, multicenter study. Infect Control Hosp Epidemiol. 2010;31:592–7. DOIPubMedGoogle Scholar
- Clincal and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 19th informational supplement, vol 29, no. 3. M100–S19. Wayne (PA): The Institute; 2009.
- Healy M, Huong J, Bittner T, Lising M, Frye S, Raza S, Microbial DNA typing by automated repetitive-sequence-based PCR. J Clin Microbiol. 2005;43:199–207. DOIPubMedGoogle Scholar
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9.PubMedGoogle Scholar
- Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37:3556–63.PubMedGoogle Scholar
- Mathema B, Mediavilla J, Kreiswirth BN. Sequence analysis of the variable number tandem repeat in Staphylococcus aureus protein A gene: spa typing. Methods Mol Biol. 2008;431:285–305.PubMedGoogle Scholar
- Chen L, Mediavilla JR, Oliveira DC, Willey BM, de Lencastre H, Kreiswirth BN. Multiplex real-time PCR for rapid staphylococcal cassette chromosome mec typing. J Clin Microbiol. 2009;47:3692–706. DOIPubMedGoogle Scholar
- Goering RV, Morrison D, Al-Doori Z, Edwards GF, Gemmell CG. Usefulness of mec-associated direct repeat unit (dru) typing in the epidemiological analysis of highly clonal methicillin-resistant Staphylococcus aureus in Scotland. Clin Microbiol Infect. 2008;14:964–9. DOIPubMedGoogle Scholar
- Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–15.PubMedGoogle Scholar
- Saïd-Salim B, Mathema B, Braughton K, Davis S, Sinsimer D, Eisner W, Differential distribution and expression of Panton-Valentine leucocidin among community-acquired methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 2005;43:3373–9. DOIPubMedGoogle Scholar
- Johnson WM, Tyler SD, Ewan EP, Ashton FE, Pollard DR, Rozee KR. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J Clin Microbiol. 1991;29:426–30.PubMedGoogle Scholar
- Diep BA, Stone GG, Basuino L, Graber CJ, Miller A, des Etages SA, The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2008;197:1523–30. DOIPubMedGoogle Scholar
- Anthony RM, Connor AM, Power EG, French GL. Use of the polymerase chain reaction for rapid detection of high-level mupirocin resistance in staphylococci. Eur J Clin Microbiol Infect Dis. 1999;18:30–4. DOIPubMedGoogle Scholar
- Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–74. DOIPubMedGoogle Scholar
- Amaral MM, Coelho LR, Flores RP, Souza RR, Silva-Carvalho MC, Teixeira LA, The predominant variant of the Brazilian epidemic clonal complex of methicillin-resistant Staphylococcus aureus has an enhanced ability to produce biofilm and to adhere to and invade airway epithelial cells. J Infect Dis. 2005;192:801–10. DOIPubMedGoogle Scholar
- Edgeworth JD, Yadegarfar G, Pathak S, Batra R, Cockfield JD, Wyncoll D, An outbreak in an intensive care unit of a strain of methicillin-resistant Staphylococcus aureus sequence type 239 associated with an increased rate of vascular access device-related bacteremia. Clin Infect Dis. 2007;44:493–501. DOIPubMedGoogle Scholar
- Cho DT, Cha HY, Chang HH, Kim SW, Chung JM, Kim J, Risk factors for specific methicillin-resistant Staphylococcus aureus clones in a Korean hospital. J Antimicrob Chemother. 2006;57:1122–7. DOIPubMedGoogle Scholar
- Li DZ, Chen YS, Yang JP, Zhang W, Hu CP, Li JS, Preliminary molecular epidemiology of the Staphylococcus aureus in lower respiratory tract infections: a multicenter study in China. Chin Med J (Engl). 2011;124:687–92.PubMedGoogle Scholar
- Smyth DS, Wong A, Robinson DA. Cross-species spread of SCCmec IV subtypes in staphylococci. Infect Genet Evol. 2011;11:446–53. DOIPubMedGoogle Scholar
- Van Bellum A, Struelens M, de Visser A, Verbrugh H, Tibayrenc M. Role of genomic typing in taxonomy, evolutionary genetics, and microbial epidemiology. Clin Microbiol Rev. 2001;14:547–60. DOIPubMedGoogle Scholar
- Olive DM, Bean P. Principles and applications of methods for DNA-based typing of microbial organisms. J Clin Microbiol. 1999;37:1661–9.PubMedGoogle Scholar
- Tumin R, Wang SH, Pancholi P, Khan Y, Hines L, Stevenson K. Relationship of social and medical factors to other antimicrobial resistance among methicillin-resistant Staphylococcus aureus strains. In: Abstracts of the 139th American Public Health Association Annual Meeting; Washington, DC, Oct 29–Nov 2, 2011. Washington (DC): American Public Health Association; 2011. Abstract 246417.
Figures
Table
Follow Up
Earning CME Credit
To obtain credit, you should first read the journal article. After reading the article, you should be able to answer the following, related, multiple-choice questions. To complete the questions (with a minimum 70% passing score) and earn continuing medical education (CME) credit, please go to www.medscape.org/journal/eid. Credit cannot be obtained for tests completed on paper, although you may use the worksheet below to keep a record of your answers. You must be a registered user on Medscape.org. If you are not registered on Medscape.org, please click on the New Users: Free Registration link on the left hand side of the website to register. Only one answer is correct for each question. Once you successfully answer all post-test questions you will be able to view and/or print your certificate. For questions regarding the content of this activity, contact the accredited provider, CME@medscape.net. For technical assistance, contact CME@webmd.net. American Medical Association’s Physician’s Recognition Award (AMA PRA) credits are accepted in the US as evidence of participation in CME activities. For further information on this award, please refer to http://www.ama-assn.org/ama/pub/category/2922.html. The AMA has determined that physicians not licensed in the US who participate in this CME activity are eligible for AMA PRA Category 1 Credits™. Through agreements that the AMA has made with agencies in some countries, AMA PRA credit may be acceptable as evidence of participation in CME activities. If you are not licensed in the US, please complete the questions online, print the certificate and present it to your national medical association for review.
Article Title: Methicillin-Resistant Staphylococcus aureus Sequence Type 239-III, Ohio, USA, 2007–2009
CME Questions
1. You are seeing a 65-year-old woman admitted with newly-diagnosed methicillin-resistant Staphylococcus aureus (MRSA) bacteremia. Which of the following strains of MRSA are most common in the United States?
A. USA 100 and MRSA ST239-III
B. USA 300 and MRSA ST239-III
C. USA 100 and USA 300
D. USA 600 and MRSA ST239-III
2. As you evaluate this patient, what should you consider regarding clinical characteristics of MRSA ST239-III in the current study?
A. Most cases of MRSA ST239-III were diagnosed in community hospitals
B. Ninety-five percent of cases of MRSA ST239-III were health care associated
C. There were no cases of MRSA ST239-III associated with implanted medical devices
D. Nearly all cases of MRSA ST239-III were bloodstream infections
3. Which of the following statements regarding the treatment and prognosis of MRSA ST239-III in the current study is most accurate?
A. There was broad susceptibility to nearly all antimicrobials tested
B. Rates of resistance to vancomycin and linezolid were approximately 90%
C. Rates of treatment failure of MRSA ST239-III were significantly higher compared with those associated with USA 100 and USA 300
D. Over 20% of cases of infection with MRSA ST239-III were fatal
4. Which of the following statements regarding molecular typing of MRSA ST239-III infections in the current study is most accurate?
A. There was a single repetitive element PCR (rep-PCR) pattern
B. There were 9 different pulsed-field gel electrophoresis (PFGE) patterns
C. Traditional PFGE testing could identify all of the bacteria subtypes
D. The cluster groupings from PFGE, rep-PCR, and dru methods were essentially identical
Activity Evaluation
|
1. The activity supported the learning objectives. |
||||
|
Strongly Disagree |
|
|
|
Strongly Agree |
|
1 |
2 |
3 |
4 |
5 |
|
2. The material was organized clearly for learning to occur. |
||||
|
Strongly Disagree |
|
|
|
Strongly Agree |
|
1 |
2 |
3 |
4 |
5 |
|
3. The content learned from this activity will impact my practice. |
||||
|
Strongly Disagree |
|
|
|
Strongly Agree |
|
1 |
2 |
3 |
4 |
5 |
|
4. The activity was presented objectively and free of commercial bias. |
||||
|
Strongly Disagree |
|
|
|
Strongly Agree |
|
1 |
2 |
3 |
4 |
5 |
1Presented in part at the 48th Annual Meeting of the Infectious Diseases Society of America, Vancouver, British Columbia, Canada, October 21–24, 2010.
Related Links
Table of Contents – Volume 18, Number 10—October 2012
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Shu-Hua Wang, The Ohio State University Wexner Medical Center, N-1120 Doan Hall, 410 West 10th Ave, Columbus, OH 43210, USA
Top