Volume 18, Number 3—March 2012
Dispatch
Escherichia coli O104:H4 Infections and International Travel
Cite This Article
Citation for Media
Abstract
We analyzed travel-associated clinical isolates of Escherichia coli O104:H4, including 1 from the 2011 German outbreak and 1 from a patient who returned from the Philippines in 2010, by genome sequencing and optical mapping. Despite extensive genomic similarity between these strains, key differences included the distribution of toxin and antimicrobial drug–resistance determinants.
In May 2011, officials in northern Germany reported a sudden surge in illness due to Shiga-toxigenic Escherichia coli (STEC). Symptoms of infection ranged from self-limiting episodes of diarrhea to life-threatening hemolytic-uremic syndrome (HUS). As of July 21, 2011, >4,075 persons in 16 countries had become ill. The outbreak was associated with an unprecedented rate of HUS (908 [22.2%] of 4,075 STEC-infected persons), and 50 persons died (1).
STEC are foodborne and waterborne pathogens. Human illness is most often associated with E. coli O157:H7, but non-O157 serogroups are also being recognized as key agents of STEC disease (2–5). The recent German outbreak was caused by E. coli O104:H4. Unlike E. coli O157:H7, which has a characteristic, sorbitol nonfermenting phenotype that is readily detected by routine laboratory testing, non-O157 E. coli strains are difficult to distinguish from the nonpathogenic E. coli strains commonly found in stool specimens, and frontline laboratories in Canada do not routinely screen for them.
This study describes 2 cases of E. coli O104:H4 infection that were imported to Canada. One case was caused by a 2011 isolate associated with the recent German outbreak. The second isolate was identified in 2010. Phenotypic and genotypic features of these 2 strains are described.
On June 1, 2011, a 67-year-old Canadian man sought treatment at an Ontario hospital with a 3-day history of bloody diarrhea. He had returned from Germany on May 27. He had no signs of HUS, and E. coli O157:H7 was not detected by routine testing. Clinical specimens from this patient were referred to the Public Health Ontario Laboratories for testing. Shiga toxin was detected by enzyme immunoassay (Meridian Biosciences, Inc., Cincinnati, OH, USA), and real–time PCR confirmed that the strain, named ON-2011, was positive for the stx2 gene and negative for the eae gene (3,6). Biochemical and serologic testing confirmed that the isolate was E. coli serogroup O104:H4. The patient recovered uneventfully (4).
Before the May 2011 outbreak in Germany, a single isolate of E. coli O104:H4 had been identified in Ontario. That isolate, ON-2010, was recovered in June 2010 from a 10-month-old boy who had returned from the Philippines 2 days earlier. He was brought to the hospital with a 1-day history of vomiting and nonbloody diarrhea. A sorbitol-nonfermenting colony was recovered from a stool specimen, but it did not react with E. coli O157 antiserum. The specimen was referred to the Canadian National Microbiology Laboratory, which confirmed E. coli O104:H4. Retrospective PCR-based testing showed that this isolate was negative for stx and eae. The infant made a full and uneventful recovery.
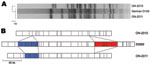
Etest (AB BIODISK, Solna, Sweden) susceptibility testing was performed by using a standard inoculum (0.5–McFarland standard); agar dilution indicated that ON-2010 was pansusceptible, whereas ON-2011 was resistant to amikacin, tetracycline, trimethoprim/sulfamethoxazole, and extended-spectrum β–lactams. PCR results for CTX–M–15 and TEM–1 were negative for ON-2010 and positive for ON-2011 (7). The gene for tellurite resistance was also absent in ON-2010, but present in ON-2011. Although both isolates were of multilocus sequence type 678, pulsed-field gel electrophoresis profiles were distinct (Figure, panel A) (8). Patterns for ON-2011 (ENXAI.0024/ENBNI.0022, PulseNet Canada designations) were identical with those reported for the current outbreak strain from Germany, whereas the ON-2010 profiles (ECXAI.2585/ECBNI.0922) were distinct (9).
Circular, highresolution NcoI restriction maps of the ON-2010 (≈5.1 Mbp) and ON-2011 (≈5.25 Mbp) genomes were generated by optical mapping (Argus Optical Mapper, OpGen, Inc, Gaithersburg, MD, USA). These were compared with an in silico map of E. coli 55989, an O104:H4 strain that was isolated in Africa and sequenced in France (10). The extensive synteny observed and the absence of large-scale genomic rearrangements (e.g., inversions, translocations) suggest that ON-2010 and ON-2011 descended from an E. coli 55989–like ancestor (Figure, panel B).
The evolutionary relationship of the 2 isolates from Ontario was further assessed by whole-genome sequencing (Roche GS–FLX Titanium; Roche Diagnostics, Laval, QC, Canada). This confirmed the genomic similarity of ON-2010 to E. coli 55989 and uncovered a 72-kb plasmid, pON-2010. Virulence genes ON-2010 and ON-2011 were compared by Bielaszewska et al. (11) (Table) on the basis of virulence factor analysis of the 2011 outbreak strain of E. coli O104:H4 from Germany. The plasmid exhibits >99% identity with p55989 and encodes the aggregative adhesion fimbriae cluster that is a defining features of enteroaggregative E. coli (EAEC) (12). In contrast, ON-2011 contains 3 plasmids. Reference-based mapping of raw sequencing reads against publicly available genome and plasmid scaffolds (HPA, BGI) confirmed that ON-2011 is a German outbreak clone (13). Except for a small number of single nucleotide polymorphisms, the clone isolated in Canada is virtually identical to those from Germany and the United Kingdom.
The O104:H4 outbreak strain from Germany exhibits key features of classic EAEC, but also contains numerous horizontally acquired virulence factors. An stx2-encoding prophage is responsible for the toxigenic properties of this strain, and multidrug-resistance loci are present, including a chromosomal tetR loci, and the plasmid-encoded CTX-M-15, TEM-1, and tellurite resistance–encoding loci (ter) (14). In contrast, the ON-2010 strain does not encode Shiga toxin, and no resistance genes have been identified. Even the tetracycline-resistance genes, independently acquired by both ON-2011 and strain 55989, are absent (Figure, panel B). However, the plasmid-encoding aggregative adhesion fimbriae cluster is present. Preliminary comparison of the 3 genomes has also shown some small-scale rearrangement events. One of these rearrangements eliminates siderophore biosynthesis genes from ON-2010 and may compromise bacterial iron acquisition. However, ON-2010 does have a strain-specific insertion that contains the hydroxyphenylacetate (hpa) operon, which may provide a nutritional advantage by enabling the organism to catabolize the phenolic and aromatic compounds abundant in the gut. This insertion is absent from the other O104:H4 genomes (15).
Since 2010, 2 cases of travel-associated E. coli O104:H4 infection have been identified in Ontario, Canada. Our analysis of these isolates could inform genomic studies on the emergence and evolution of the O104 clone first observed in Europe. The relationship of these strains was assessed by a combination of traditional, molecular, and genomic approaches. Optical mapping and sequencing indicate that ON-2010 and ON-2011 exhibit extensive synteny and are derived from a common EAEC ancestor. However, a series of horizontal gene transfer events has contributed to genotypic and phenotypic divergence. The outbreak clone from Germany, ON-2011, is an antimicrobial drug–resistant STEC isolate, whereas ON-2010 is pansusceptible and nontoxigenic.
These differences are clinically important but are not detected by diagnostic strategies that use serotype as a proxy for pathogenic capacity. Given the potential for severe clinical sequelae of STEC infections, clinical testing should use improved and affordable methods for identification of Shiga toxin that can be easily integrated into routine clinical microbiology laboratories (4,9). Future capacity to detect additional virulence factors, such as aggregative adhesion fimbriae, would enable expanded diagnostic capacity to detect pathogenic E. coli subtypes that cause human disease.
Dr Alexander is a research scientist at the Public Health Ontario Laboratories. His research interests include the application of molecular epidemiology and genomic tools to the surveillance of infectious disease and study of emerging pathogens.
Acknowledgment
We thank the Public Health Agency of Canada/Public Health Ontario E. coli O104 Research Group; the clinical and research staff at the Public Health Ontario Laboratories who were involved in the investigation of STEC; Linda Chui for her advice and support; and National Microbiology Laboratory personnel, including Celine Nadon and Michael Mulvey, for their contributions and expertise.
References
- World Health Organization. Outbreaks of E. coli O104:H4 infection: update 30. 2011 [cited 2011 July 29]. http://www.euro.who.int/en/what-we-do/health-topics/emergencies/international-health-regulations/news/news/2011/07/outbreaks-of-e.-coli-o104h4-infection-update-30
- Brooks JT, Sowers EG, Wells JG, Greene KD, Griffin PM, Hoekstra RM, Non-O157 Shiga toxin–producing Escherichia coli infections in the United States, 1983–2002. J Infect Dis. 2005;192:1422–9. DOIPubMedGoogle Scholar
- Couturier MR, Lee B, Zelyas N, Chui L. Shiga-toxigenic Escherichia coli detection in stool samples screened for viral gastroenteritis in Alberta, Canada. J Clin Microbiol. 2011;49:574–8. DOIPubMedGoogle Scholar
- Gould LH, Bopp C, Strockbine N, Atkinson R, Baselski V, Body B, Recommendations for diagnosis of Shiga toxin–producing Escherichia coli infections by clinical laboratories. MMWR Recomm Rep. 2009;58(RR-12):1–14.PubMedGoogle Scholar
- Hadler JL, Clogher P, Hurd S, Phan Q, Mandour M, Bemis K, Ten-year trends and risk factors for non-O157 Shiga toxin–producing Escherichia coli found through Shiga toxin testing, Connecticut, 2000–2009. Clin Infect Dis. 2011;53:269–76. DOIPubMedGoogle Scholar
- Chui L, Couturier MR, Chiu T, Wang G, Olson AB, McDonald RR, Comparison of Shiga toxin-producing Escherichia coli detection methods using clinical stool samples. J Mol Diagn. 2010;12:469–75. DOIPubMedGoogle Scholar
- Pitout JD, Hamilton N, Church DL, Nordmann P, Poirel L. Development and clinical validation of a molecular diagnostic assay to detect CTX-M-type β-lactamases in Enterobacteriaceae. Clin Microbiol Infect. 2007;13:291–7. DOIPubMedGoogle Scholar
- Ribot EM, Fair M, Gautom R, Cameron D, Hunter S, Swaminathan B, Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157: H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. One-day (24–28 h) standardized laboratory protocol for molecular subtyping of Escherichia coli O157:H7, non-typhoidal Salmonella serotypes, and Shigella sonnei by pulsed-field gel electrophoresis (PFGE). Atlanta (GA): US Department of Health and Human Services; 2007.
- Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. DOIPubMedGoogle Scholar
- Bielaszewska M, Mellmann A, Zhang W, Kock R, Fruth A, Bauwens A, Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect Dis. 2011;11:671–6.PubMedGoogle Scholar
- Bernier C, Gounon P, Le Bouguenec C. Identification of an aggregative adhesion fimbria (AAF) type III–encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect Immun. 2002;70:4302–11. DOIPubMedGoogle Scholar
- Health Protection Agency. LGP bioinformatics portal. 2011 [cited 2011 Jul 29]. http://www.hpa-bioinformatics.org.uk/lgp/genomes
- Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011;365:709–17. DOIPubMedGoogle Scholar
- Smith EA, Macfarlane GT. Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Bacteriol. 1996;81:288–302. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleTable of Contents – Volume 18, Number 3—March 2012
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Vanessa G. Allen, Public Health Ontario–Public Health Laboratories, 81 Resources Rd, Toronto, ON M9P 3T1, Canada
Top