Volume 18, Number 4—April 2012
Dispatch
Characterization of Mycobacterium orygis as M. tuberculosis Complex Subspecies
Figure 1
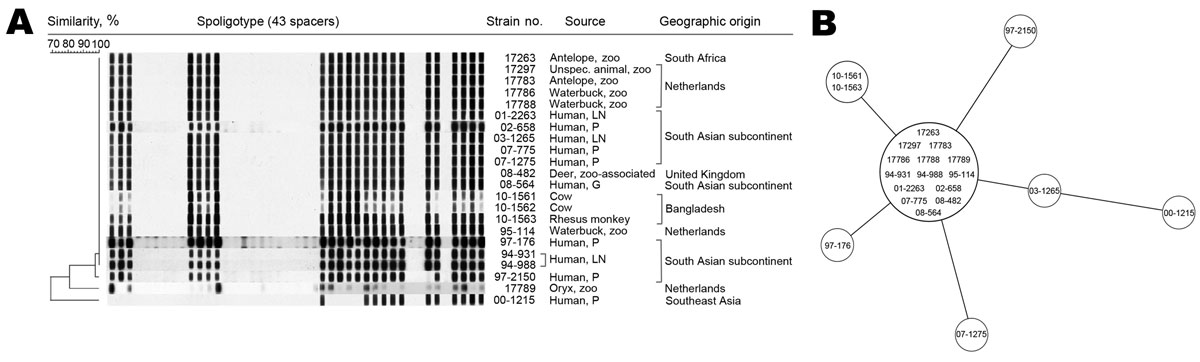
Figure 1. Spoligotyping and 24-locus variable number tandem repeat (VNTR) typing results for Mycobacterium orygis. A) Spoligotyping patterns for the oryx bacillus isolates in this study; ST587 is the most common pattern (labeled SB0422 at www.mbovis.org), with minor deviations. B) Minimum spanning tree based on 24-locus VNTR typing results for the oryx bacillus isolates in this study. One type dominates, with few strains representing minor variations. The dominant clone includes isolates from humans and animals. P, pulmonary; LN, lymph node; G, gastric juice. Both panels were created by using BioNumerics version 6.1 software (Applied Maths, Sint-Martens-Latem, Belgium); similarity coefficients were calculated by using Dice (spoligotyping) and Pearson (VNTR) methods; cluster analysis was done by the UPGMA (unweighted pair group method with arithmetic mean). Isolate 10–1562 (cow, Bangladesh) could not be included in (B) because of insufficient DNA.