Volume 18, Number 4—April 2012
Research
Geographic Distribution of Hantaviruses Associated with Neotomine and Sigmodontine Rodents, Mexico
Figure 3
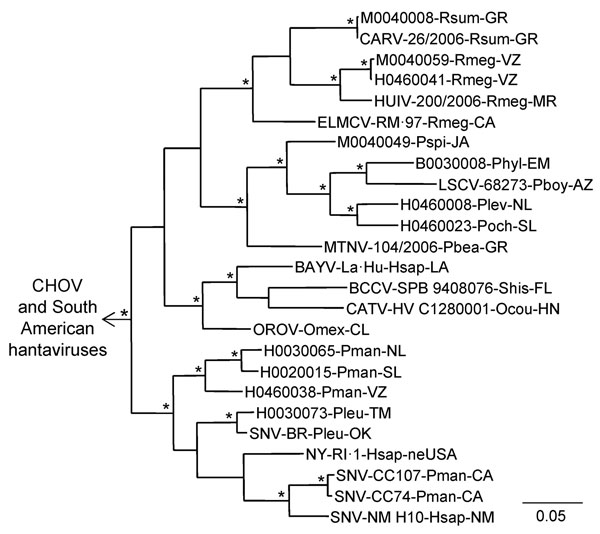
Figure 3. Results of the Bayesian analyses of the nucleotide sequences of a 1,078-nt fragment of the glycoprotein precursor genes of 11 of the 24 hantaviruses found in Mexico in this study and 20 other hantaviruses naturally associated with members of the Neotominae or Sigmodontinae. An asterisk at a node indicates that the probability values in support of the clade were >0.95. Scale bar indicates substitutions per site. The branch labels include (in the following order) virus, strain, host species, and state or country. BAYV, GenBank accession no. L36930; BCCV, L39950; CARV, AB620104; CATV, DQ177347; CHOV, DQ285047; ELMCV, U26828; HUIV, AB620107; LSCV, AF307323; MTNV, AB620101; NYV, U36801; OROV, EF534080; SNV—strains BR (AF030552), CC74 (L33684), CC107 (L33474), and NM H10 (L25783). The viruses from South America were Andes virus, GenBank accession no. AF291703; Caño Delgadito virus, DQ284451; Laguna Negra virus, AF005728; Maporal virus, AY363179; and Rio Mamoré virus, FJ608550. The designated outgroup was Andes virus strain Chile-9717869.
1These authors contributed equally to this article.