Volume 18, Number 4—April 2012
Research
Geographic Distribution of Hantaviruses Associated with Neotomine and Sigmodontine Rodents, Mexico
Cite This Article
Citation for Media
Abstract
To increase our knowledge of the geographic distribution of hantaviruses associated with neotomine or sigmodontine rodents in Mexico, we tested 876 cricetid rodents captured in 18 Mexican states (representing at least 44 species in the subfamily Neotominae and 10 species in the subfamily Sigmodontinae) for anti-hantavirus IgG. We found antibodies against hantavirus in 35 (4.0%) rodents. Nucleotide sequence data from 5 antibody-positive rodents indicated that Sin Nombre virus (the major cause of hantavirus pulmonary syndrome [HPS] in the United States) is enzootic in the Mexican states of Nuevo León, San Luis Potosí, Tamaulipas, and Veracruz. However, HPS has not been reported from these states, which suggests that in northeastern Mexico, HPS has been confused with other rapidly progressive, life-threatening respiratory diseases. Analyses of nucleotide sequence data from 19 other antibody-positive rodents indicated that El Moro Canyon virus and Limestone Canyon virus are geographically widely distributed in Mexico.
Hantavirus pulmonary syndrome (HPS) is a potentially fatal zoonosis caused by hantaviruses (family Bunyaviridae, genus Hantavirus) that are principally associated with members of the rodent family Cricetidae, more specifically, members of the subfamily Neotominae or Sigmodontinae (1,2). The viruses known to cause HPS on the North American continent are Bayou virus, Black Creek Canal virus (BCCV), Choclo virus (CHOV), New York virus, and Sin Nombre virus (SNV) (3–7). Other hantaviruses that are principally associated with neotomine or North American sigmodontine rodents include Carrizal virus (CARV), Catacamas virus, El Moro Canyon virus (ELMCV), Huitzilac virus (HUIV), Limestone Canyon virus (LSCV), Montano virus (MTNV), Muleshoe virus (MULV), Playa de Oro virus, and Rio Segundo virus (RIOSV) (8–14).
Specific rodents (usually 1 or 2 closely related species) are the principal hosts of the hantaviruses, for which natural host relationships have been well characterized. The current principal host relationships of some hantaviruses seem to represent a long-term association between viruses in the genus Hantavirus and rodents in the family Cricetidae. Evidence for this ancient relationship includes the association of phylogenetically closely related hantavirus species with phylogenetically closely related allopatric rodent species. For example, Catacamas virus is associated with Coues’s rice rat (Oryzomys couesi) in Honduras, and Bayou virus is associated with the marsh rice rat (Oryzomys palustris) in the southeastern United States (9,15).
The rodent fauna of Mexico comprises the brush mouse (Peromyscus boylii), the deer mouse (P. maniculatus), the western harvest mouse (Reithrodontomys megalotis), the hispid cotton rat (Sigmodon hispidus), the fulvous pygmy rice rat (Oligoryzomys fulvescens), and 122 other species in the Neotominae or Sigmodontinae (16). In the southwestern United States, LSCV, SNV, ELMCV, and MULV are principally associated with rodents of the species P. boylii, P. maniculatus, R. megalotis, and S. hispidus, respectively (10–12,17), and that in Panama, CHOV is principally associated with O. fulvescens (18). Hypothetically, LSCV, SNV, ELMCV, and/or MULV—in association with deer mice (Peromyscus spp.), harvest mice (Reithrodontomys spp.), or cotton rats (Sigmodon spp.)—are widely distributed in northern Mexico, and the hantavirus assemblage of southern Mexico includes CHOV or hantaviruses that are phylogenetically closely related to CHOV.
Our knowledge of the rodent-associated hantaviruses in Mexico includes the following findings: HUIV RNA in a western harvest mouse (R. megalotis) captured in Morelos (8); CARV RNA in a Sumichrast’s harvest mouse (R. sumichrasti) and MTNV RNA in an Orizaba deer mouse (P. beatae) from Guerrero (8); Playa de Oro virus RNA in a Mexican oryzomys (Oryzomys mexicanus) and Jaliscan cotton rat (S. mascotensis) from Colima (13); ELMCV RNA and SNV RNA in western harvest mice from Zacatecas (14); antibody against hantavirus in nimble-footed mice (P. levipes) captured in Tamaulipas (19); and antibody against hantavirus in a North American deer mouse (P. maniculatus), transvolcanic mice (P. hylocetes), black-eared mice (P. melanotis), and Sumichrast’s harvest mouse captured in the state of Mexico (20,21). The purpose of this study was to extend our knowledge of the geographic distribution of hantaviruses associated with neotomine or sigmodontine rodents in Mexico.
Blood samples from 876 rodents, representing at least 44 species in the Neotominae and 10 species in the Sigmodontinae, were tested for anti-hantavirus IgG. The 876 rodents were captured during 1998–2008 at 43 localities in 18 states in Mexico (Table A1). Blood samples from all of the rodents and lung samples from the antibody-positive rodents were acquired from the Natural Science Research Laboratory, Museum of Texas Tech University, Lubbock, Texas, USA.
The blood samples were tested for IgG to Caño Delgadito virus (CADV) strain VHV-574 by using an ELISA in which CADV can be highly cross-reactive with SNV, BCCV, and other North American hantaviruses (22). The antibody titers in the antibody-positive blood samples were recorded as 320, 1,280, or >5,120.
Samples of lung tissue from the antibody-positive rodents were tested for hantavirus nucleocapsid (N) protein gene RNA. Subsequently, we determined the nucleotide sequences of a 1,078-nt fragment of the glycoprotein precursor (GPC) genes of 11 of the hantaviruses associated with the antibody-positive rodents. We chose these 11 viruses to represent the geographic distribution and natural host associations of the hantaviruses in Mexico included in this study. Total RNA was isolated from 30 mg to 45 mg of lung tissue by using Tri Reagent (Sigma-Aldrich, St. Louis, MO, USA). First-strand cDNA was synthesized from small (S) segment and medium (M) segment RNA by using SuperScript II RNase H– Reverse Transcriptase (Invitrogen Life Technologies, Inc., Carlsbad, CA, USA) in conjunction with oligonucleotide 5′-GGTGGTTGTGGTAGTAGTAGACTCC-3′ (23). The first-round and second-round (hemi-nested) PCR assays used the MasterTaq Kit (Eppendorf North America, Inc., Westbury, NY, USA). (The sequences of the oligonucleotides that were used to prime the PCR are available from the corresponding author.) The sizes of the N protein gene amplicons from the second-round assays ranged from 377 to 545 bp, the sizes of the GPC gene amplicons from the second-round assays ranged from 607 to 631 bp (M1 amplicon) and 571 to 618 bp (M2 amplicon), and the lengths of the overlaps between the M1 and M2 amplicons ranged from 125 to 134 bp. Together, the nucleotide sequences of the M1 and M2 amplicons encoded a 359-aa fragment of the GC glycoprotein.
The sequences in each dataset were aligned by using the computer program ClustalW version 2.0.12 (24). Sequence nonidentities were equivalent to uncorrected (p) distances. The phylogenetic analyses of nucleotide sequences were conducted with MRBAYES 3.1.2 (25) and programs in the computer software package PAUP* (26). The Bayesian analyses used the general time reversible + proportion invariant + Γ model and the following options in MRBAYES 3.1.2: two simultaneous runs of 4 Markov chains, 2 million generations, and sample frequency = every 1,000th generation. The first 1,000 trees were discarded after review of the likelihood scores, convergence statistics, and potential scale reduction factors; and a consensus tree (50% majority rule) was constructed from the remaining trees. Probability values in support of the clades were calculated a posteriori, and clades with probability values >0.95 were considered supported by the data (27).
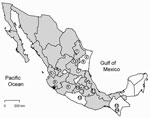
Antibody (IgG) against hantavirus was found in 35 (4.0%) of 876 rodents captured in 18 states in Mexico (Table A1). The antibody-positive rodents were from 16 localities (Table A2) in 14 municipalities in 10 states: Chiapas, Guerrero, Jalisco, México, Michoacán, Nayarit, Nuevo León, San Luis Potosí, Tamaulipas, and Veracruz (Figure 1). None of the rodents captured in Chihuahua (n = 9), Coahuila (n = 16), Guanajuato (n = 8), Oaxaca (n = 64), Puebla (n = 15), Sinaloa (n = 9), Sonora (n = 22), or Tlaxcala (n = 16) were antibody-positive to CADV strain VHV-574.
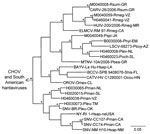
Hantavirus N protein gene RNA was detected in samples of lung from 24 (68.6%) of the 35 antibody-positive rodents (Table). The Bayesian analyses of the N protein gene sequences separated the 24 Mexican viruses in this study into 4 groups (Figure 2). Group I included CARV, group 2 included HUIV, group III included LSCV and MTNV, and group IV included SNV strains Convict Creek 74, Convict Creek 107, and NM H10.
Hantavirus GPC gene RNA was detected in each of the 11 rodents assayed for GPC gene RNA (Table). The topology of the GPC gene tree (Figure 3) was essentially identical to the topology of the N protein gene tree (Figure 2) with respect to relationships between the viruses from Mexico in this study, CARV, HUIV, MTNV, and the other hantaviruses found in North America. M0040008, CARV, M0040059, H0460041, HUIV, and ELMCV were monophyletic in the Bayesian analyses of the GPC gene sequence data (Figure 3) and N protein gene sequence data (Figure 2).
Nonidentities among the amino acid sequences of the 359-aa fragment of the GC glycoproteins of the 11 hantaviruses from Mexico in this study, CARV, HUIV, and MTNV ranged from 0% to 18.4% (Table A3). Nonidentities between the sequences of the 359-aa fragment of the GC glycoproteins of these 14 hantaviruses and the sequences of the homologous fragment of the GC glycoproteins of the other hantaviruses found in North America ranged from 1.1% (H0030073 and SNV strain Blue River-Oklahoma) to 18.9% (M0040049 and BCCV strain SPB 9408076).
The Eighth Report of the International Committee on Taxonomy of Viruses sets forth the criteria for species demarcation in the genus Hantavirus (1). One of these criteria is that strains of different species must exhibit at least a 7% difference in amino acid sequence identity in comparisons of complete N protein sequences and in comparisons of complete GPC sequences.
ELMCV was first described in 1994 (10); LSCV was described in 2001 (11); and CARV, HUIV, and MTNV were described in 2011 (8). In a previous study (8), the amino acid sequence of the N protein of MTNV was 5.8% different from the amino acid sequence of the N protein of LSCV strain 68273, and the amino acid sequence of the N protein of CARV was 3.7% different from the amino acid sequence of the N protein of ELMCV strain RM•97 and 8.4% different from the amino acid sequence of the N protein of RIOSV strain RMx•Costa•1. The amino acid sequence of the N protein of HUIV was 1.4% different from the amino acid sequence of the N protein of ELMCV strain RM•97 and 8.4% different from the amino acid sequence of the N protein of RIOSV strain RMx•Costa•1. Accordingly, MTNV should be considered a strain of LSCV, and CARV and HUIV could be considered strains of ELMCV or RIOSV. Alternatively, CARV and HUIV could be considered members of a species complex that includes ELMCV, RIOSV, and other hantaviruses that are naturally associated with harvest mice (Reithrodontomys spp.). There is presumptive evidence for RIOSV or hantavirus(es) that are genetically closely related to RIOSV in Sumichrast’s harvest mice, a Mexican harvest mouse (R. mexicanus), and a Chiriqui harvest mouse (R. creper) captured in Panama (29).
Collectively, the results of the Bayesian analyses of N protein gene sequence data (Figure 2), Bayesian analyses of the GPC gene sequence data (Figure 3), and pairwise comparisons of GC sequences (Table A3) indicate that H0020015, H0030065, H0030073, and H0460038 are strains of SNV. The results of these analyses also indicate that M0040008, M0040059, and H0460041 are strains of ELMCV or RIOSV and that B0030008, H0460008, H0460023, and M0040049 are strains of LSCV. Accordingly, the N protein gene RNA–positive rodents in this study (Table) were infected with SNV, ELMCV, RIOSV, or LSCV.
Specific knowledge of the natural host relationships of LSCV previously was limited to LSCV RNA in 6 brush mice (P. boylii) captured in northern Arizona (11). The results of this study indicate that the transvolcanic mouse (P. hylocetes) and the black-eared mouse (P. melanotis) in the state of México, the nimble-footed mouse (P. levipes) in Nuevo León, the El Carrizo deer mouse (P. ochraventer) in San Luis Potosí, and the gleaning mouse (P. spicilegus) in Jalisco are natural but not necessarily principal hosts of LSCV.
HPS was first recognized as a distinct clinical entity in the southwestern United States in 1993 (30). Through 2009, a total of 510 HPS cases were reported to the National Notifiable Diseases Surveillance System or registered by the Centers for Disease Control and Prevention (31). Most of these cases occurred in the southwestern United States, 92 (33.7%) of 273 HPS cases that occurred in the southwestern United States before 2010 were fatal, and all of the cases from the southwestern United States through 2009 were attributed to SNV.
The results of this study indicate that SNV is widely distributed in northeastern Mexico. The geographic distribution of deer mice (P. maniculatus) in Mexico includes 23 states (28), and ≈20 million persons lived in rural areas in this 23-state region in 2010 (32). Yet, to our knowledge, no cases of HPS have been reported from northeastern Mexico or elsewhere in Mexico.
We hypothesize that HPS caused by SNV in Mexico has been confused with other rapidly progressive, life-threatening respiratory diseases (e.g., plague, tularemia, pneumococcal pneumonia, influenza). Alternatively, SNV in Mexico is substantially less virulent than SNV in the western United States, or human contact with SNV-infected rodents in Mexico is less frequent or less intimate than human contact with SNV-infected rodents in the western United States.
Laboratory confirmation of the diagnoses of most HPS cases in the United States before 2010 was based on the results of serologic assays in which ELMCV and LSCV can be highly cross-reactive with SNV (31). Thus, in all likelihood, some of the HPS cases from the western United States were actually caused by ELMCV or LSCV, and these viruses as well as SNV are etiologic agents of HPS in Mexico.
Ms Milazzo is a senior research associate at the University of Texas Medical Branch, Galveston. Her scientific interests include the epidemiology and ecology of New World rodent-borne RNA viruses.
Acknowledgments
We thank Robert J. Baker for facilitating the loan of the samples from the Natural Science Research Laboratory, Museum of Texas Tech University; Eleanor R. Deardorff and Avery O. Tatters for assisting with the genetic characterization of the hantaviruses in Mexico.
This study was financially supported by National Institutes of Health grants AI-041435 and AI-067947. Christina Cassetti and Patricia Repik facilitated the grant support for this study.
References
- Nichol ST, Beaty BJ, Elliott RM, Goldbach R, Plyusnin A, Schmaljohn CS, Family Bunyaviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. San Diego (CA): Elsevier Academic Press; 2005, p. 695–716.
- Musser GG, Carleton MD. Family Cricetidae. In: Wilson DE, Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference, 3rd ed. Baltimore: Johns Hopkins University Press; 2005. p. 955–1189.
- Khan AS, Spiropoulou CF, Morzunov S, Zaki SR, Kohn MA, Nawas SR, Fatal illness associated with a new hantavirus in Louisiana. J Med Virol. 1995;46:281–6. DOIPubMedGoogle Scholar
- Khan AS, Gaviria M, Rollin PE, Hlady WG, Ksiazek TG, Armstrong LR, Hantavirus pulmonary syndrome in Florida: association with the newly identified Black Creek Canal virus. Am J Med. 1996;100:46–8. DOIPubMedGoogle Scholar
- Nelson R, Cañate R, Pascale JM, Dragoo JW, Armien B, Armien AG, Confirmation of Choclo virus as the cause of hantavirus cardiopulmonary syndrome and high serum antibody prevalence in Panama. J Med Virol. 2010;82:1586–93. DOIPubMedGoogle Scholar
- Hjelle B, Lee SW, Song W, Torrez-Martinez N, Song JW, Yanagihara R, Molecular linkage of hantavirus pulmonary syndrome to the white-footed mouse, Peromyscus leucopus: genetic characterization of the M genome of New York virus. J Virol. 1995;69:8137–41.PubMedGoogle Scholar
- Ksiazek TG, Peters CJ, Rollin PE, Zaki S, Nichol S, Spiropoulou C, Identification of a new North American hantavirus that causes acute pulmonary insufficiency. Am J Trop Med Hyg. 1995;52:117–23.PubMedGoogle Scholar
- Kariwa H, Yoshida H, Sánchez-Hernández C, Romero-Almaraz MD, Almazán-Catalán JA, Ramos C, Genetic diversity of hantaviruses in Mexico: identification of three novel hantaviruses from Neotominae rodents. Virus Res. 2011; Epub ahead of print.PubMedGoogle Scholar
- Milazzo ML, Cajimat MNB, Hanson JD, Bradley RD, Quintana M, Sherman C, Catacamas virus, a hantaviral species naturally associated with Oryzomys couesi (Coues’ oryzomys) in Honduras. Am J Trop Med Hyg. 2006;75:1003–10.PubMedGoogle Scholar
- Hjelle B, Chavez-Giles F, Torrez-Martinez N, Yates T, Sarisky J, Webb J, Genetic identification of a novel hantavirus of the harvest mouse Reithrodontomys megalotis. J Virol. 1994;68:6751–4.PubMedGoogle Scholar
- Sanchez AJ, Abbott KD, Nichol ST. Genetic identification and characterization of Limestone Canyon virus, a unique Peromyscus-borne hantavirus. Virology. 2001;286:345–53. DOIPubMedGoogle Scholar
- Rawlings JA, Torrez-Martinez N, Neill SU, Moore GM, Hicks BN, Pichuantes S, Cocirculation of multiple hantaviruses in Texas, with characterization of the small (S) genome of a previously undescribed virus of cotton rats (Sigmodon hispidus). Am J Trop Med Hyg. 1996;55:672–9.PubMedGoogle Scholar
- Chu Y-K, Owen RD, Sánchez-Hernández C, Romero-Almaraz ML, Jonsson CB. Genetic characterization and phylogeny of a hantavirus from western Mexico. Virus Res. 2008;131:180–8. DOIPubMedGoogle Scholar
- Hjelle B, Anderson B, Torrez-Martinez N, Song W, Gannon WL, Yates TL. Prevalence and geographic genetic variation of hantaviruses of New World harvest mice (Reithrodontomys): identification of a divergent genotype from a Costa Rican Reithrodontomys mexicanus. Virology. 1995;207:452–9. DOIPubMedGoogle Scholar
- Ksiazek TG, Nichol ST, Mills JN, Groves MG, Wozniak A, McAdams S, Isolation, genetic diversity, and geographic distribution of Bayou virus (Bunyaviridae: Hantavirus). Am J Trop Med Hyg. 1997;57:445–8.PubMedGoogle Scholar
- Ceballos G, Arroyo-Cabrales J, Medellín RA. Lista sistemática de las especies. In: Ceballos G, Oliva G, editors. Los mamíferos silvestres de México. Mexico City: Comisión Nacional para el Conocimiento y Uso de la Biodiversidad Fondo de Cultura Económica; 2005. p. 73–95.
- Childs JE, Ksiazek TG, Spiropoulou CF, Krebs JW, Morzunov S, Maupin GO, Serologic and genetic identification of Peromyscus maniculatus as the primary rodent reservoir for a new hantavirus in the southwestern United States. J Infect Dis. 1994;169:1271–80. DOIPubMedGoogle Scholar
- Vincent MJ, Quiroz E, Gracia F, Sanchez AJ, Ksiazek TG, Kitsutani PT, Hantavirus pulmonary syndrome in Panama: identification of novel hantaviruses and their likely reservoirs. Virology. 2000;277:14–9. DOIPubMedGoogle Scholar
- Castro-Arellano I, Suzán G, León RF, Jiménez RM, Lacher TE Jr. Survey for antibody to hantaviruses in Tamaulipas, México. J Wildl Dis. 2009;45:207–12.PubMedGoogle Scholar
- Mantooth SJ, Milazzo ML, Bradley RD, Hice CL, Ceballos G, Tesh RB, Geographical distribution of rodent-associated hantaviruses in Texas. J Vector Ecol. 2001;26:7–14.PubMedGoogle Scholar
- Suzán G, Ceballos G, Mills J, Ksiazek TG, Yates T. Serologic evidence of hantavirus infection in sigmodontine rodents in Mexico. J Wildl Dis. 2001;37:391–3.PubMedGoogle Scholar
- Fulhorst CF, Monroe MC, Salas RA, Duno G, Utrera A, Ksiazek TG, Isolation, characterization and geographic distribution of Caño Delgadito virus, a newly discovered South American hantavirus (family Bunyaviridae). Virus Res. 1997;51:159–71. DOIPubMedGoogle Scholar
- Morzunov SP, Feldmann H, Spiropoulou CF, Semenova VA, Rollin PE, Ksiazek TG, A newly recognized virus associated with a fatal case of hantavirus pulmonary syndrome in Louisiana. J Virol. 1995;69:1980–3.PubMedGoogle Scholar
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W (1.7): improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choices. Nucleic Acids Res. 1994;22:4673–80. DOIPubMedGoogle Scholar
- Huelsenbeck JP, Ronquist FR. MRBAYES: Bayesian inference of phylogenic trees. Bioinformatics. 2001;17:754–5. DOIPubMedGoogle Scholar
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sunderland (MA): Sinauer Associates, Inc.; 2002.
- Erixon P, Svennblad B, Britton T, Oxelman B. Reliability of Bayesian posterior probabilities and bootstrap frequencies in phylogenetics. Syst Biol. 2003;52:665–73. DOIPubMedGoogle Scholar
- Ramírez-Pulido J, Sánchez A, Aguilera U, Castro-Campillo A. Peromyscus maniculatus. In: Ceballos G, Oliva G, editors. Los Mamiferos Silvestres de Mexico. Mexico City: Comisión Nacional para el Conocimiento y Uso de la Biodiversidad Fondo de Cultura Económica; 2005, p. 748–50.
- Salazar-Bravo J, Armién B, Suzán G, Armién A, Ruedas LA, Avila M, Serosurvey of wild rodents for hantaviruses in Panama, 2000–2002. J Wildl Dis. 2004;40:103–9.PubMedGoogle Scholar
- Duchin JS, Koster FT, Peters CJ, Simpson GL, Tempest B, Zaki SR, Hantavirus pulmonary syndrome: a clinical description of 17 patients with a newly recognized disease. N Engl J Med. 1994;330:949–55. DOIPubMedGoogle Scholar
- MacNeil A, Ksiazek TG, Rollin PE. Hantavirus pulmonary syndrome, United States, 1993–2009. Emerg Infect Dis. 2011;17:1195–201. DOIPubMedGoogle Scholar
- Anuario estadístico de los Estados Unidos Mexicanos, 2010. Instituto Nacional de Estadística y Geografía [cited 2011 Jul 8]. http://www.inegi.org.mx
Figures
Tables
Cite This Article1These authors contributed equally to this article.
Table of Contents – Volume 18, Number 4—April 2012
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Charles F. Fulhorst, The University of Texas Medical Branch, 301 University Blvd, Galveston, TX 77555-0609, USA
Top