Volume 19, Number 11—November 2013
Dispatch
Tula Hantavirus Infection in Immunocompromised Host, Czech Republic
Figure 1
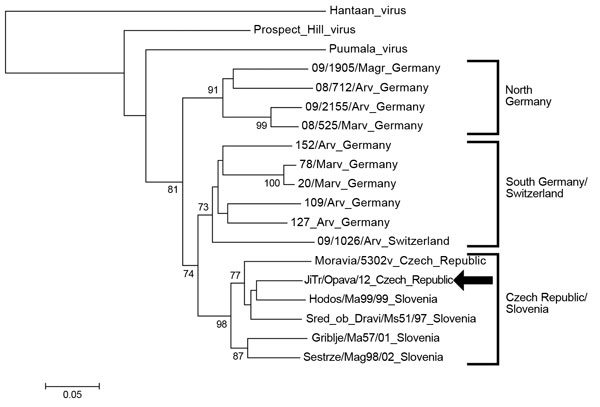
Figure 1. . . Phylogenetic tree (neighbor-joining analysis with maximum composite likelihood method) of Tula virus on the basis of large segment partial sequences (nt 2957–3337), Ostrava, Czech Republic, October 2012 GenBank accesion numbers: Haantaan virus (NC_005222), Puumala virus (Z66548), Prospect Hill virus (EF646763), 09/1905/Magr (HQ728460), 08/712/Arv (HQ728453), 09/2155/Arv (HQ728456), 08/525/Marv (HQ728461), 152/Arv (HQ728459), 78/Marv (HQ728464), 20/Marv (HQ728462), 109/Arv (HQ728457), 127/Arv (HQ728458), 09/1026/Arv (HQ728455), Moravia/5302v (AJ005637), JiTr/Opava /12 (KC522413), Hodos/Ma99/99 (FJ495101), Sred ob Dravi/Ms51/97 (FJ495102), Griblje/Ma57/01 (FJ495099), Sestrze/Mag98/02 (FJ495100). Bootstrap values ≥70%, calculated from 1,000 replicates, are shown at the tree branches. Arrow indicates strain isolated in this study. The tree is drawn to scale. The scale bar indicates an evolutionary distance of 0.05 substitutions per position in the sequence.
1The authors contributed equally to this article.