Phylogenetic and Ecologic Perspectives of a Monkeypox Outbreak, Southern Sudan, 2005
Yoshinori Nakazawa

, Ginny L. Emerson, Darin S. Carroll, Hui Zhao, Yu Li, Mary G. Reynolds, Kevin L. Karem, Victoria A. Olson, R. Ryan Lash, Whitni B. Davidson, Scott K. Smith, Rebecca S. Levine, Russell L. Regnery, Scott A. Sammons, Michael A. Frace, Elmangory M. Mutasim, Mubarak E. M. Karsani, Mohammed O. Muntasir, Alimagboul A. Babiker, Langova Opoka, Vipul Chowdhary, and Inger K. Damon
Author affiliations: Author affiliations: Centers for Disease Control and Prevention, Atlanta, Georgia, USA (Y. Nakazawa, G. L. Emerson, D.S. Carroll, H. Zhao, Y. Li, M.G. Reynolds, K. L. Karem, V. A. Olson, R. R. Lash, W. B. Davidson, S. K. Smith, R. S. Levine, R. L. Regnery, S. A. Sammons, M.A. Frace, I.K. Damon); National Public Health Laboratory, Khartoum, Sudan (E.M. Mutasim, M.E.M. Karsani); Federal Ministry of Health, Khartoum (M.O. Muntasir, A.A. Babiker); World Health Organization Regional Office for the Eastern Mediterranean, Cairo, Egypt (M.L. Opoka); Médecins Sans Frontières, Khartoum (V. Chowdhary)
Main Article
Figure 3
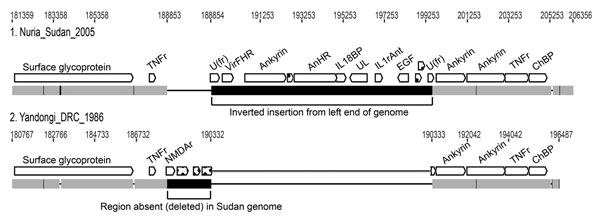
Figure 3. . Comparison of a right-end segment from genomes of monkeypox virus (Nuria Sudan 2005 and Yandongi DRC1986. Numbers above genome map are nucleotide positions. Gray boxes represent DNA sequence identity in the 2 genomes; black represents differences. The 2 large black boxes illustrate the insertion/deletion event found in Sudan isolates 1 and 2. A region from the left end of the genome has been inserted where a portion of the right end (shown in Yandongi) has been deleted. Thin black horizontal lines represent gaps in the alignment. Open reading frames are shown in white. Open reading frame names were assigned with reference to MPXV genomes available at the Poxvirus Bioinformatics Resource Center (www.poxvirus.org). Segment boxes with dots indicate unknown genome sequences; TNFr, tumor necrosis factor receptor; U(fr), unknown fragment; VirFHR, virulence factor host range; AnHR, ankyrin host range; IL18BP, interleukin 18 binding protein; UL, ubiquitin ligase; IL1rAnt interleukin 1 receptor antagonist; EGF, epidermal growth factor; ChBP, chemokine binding protein; NMDAr, N-methyl D-aspartate receptor-like protein.
Main Article
Page created: January 22, 2013
Page updated: January 22, 2013
Page reviewed: January 22, 2013
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
