Volume 19, Number 5—May 2013
Research
Targeting Surveillance for Zoonotic Virus Discovery
Abstract
We analyzed a database of mammal–virus associations to ask whether surveillance targeting diseased animals is the best strategy to identify potentially zoonotic pathogens. Although a mixed healthy and diseased animal surveillance strategy is generally best, surveillance of apparently healthy animals would likely maximize zoonotic virus discovery potential for bats and rodents.
Nearly two thirds of emerging infectious diseases that affect humans are zoonotic, and three fourths of these originate in wildlife, making surveillance of wildlife for novel pathogens part of a logical strategy to prevent the future emergence of zoonoses (1–4). Wildlife are thought to harbor a high diversity of unknown pathogens, but global characterization of this diversity would be costly and logistically challenging (5). Given limited resources for pandemic prevention, there is public health benefit in focusing pathogen discovery on those species most likely to harbor novel zoonoses (3,4).
One strategy to maximize the likelihood of discovering novel pathogens is surveillance of animal die-offs, outbreaks in wildlife, or diseased wildlife. We analyzed a database of known zoonotic viruses in mammal hosts to answer the driving question of whether we should stratify surveillance strategies (i.e., conduct surveillance of visibly diseased vs. apparently healthy animals) by wildlife host groups to best detect novel pathogens with zoonotic potential. In answering this question, we can better determine how host and virus taxonomy might influence our decisions about applying limited surveillance resources to a growing global health problem.
We focused our analysis on mammalian hosts and viruses because they, more than any other host–pathogen type, are likely to be associated with emerging infectious diseases of humans (3,6). We constructed a database of all emerging viruses of humans that were previously identified as originating in wildlife; the database was supplemented with all zoonotic viruses with nonhuman mammalian hosts found in the International Committee on the Taxonomy of Viruses database (www.ictvdb.org) (2). For each zoonotic virus, we conducted a literature search for reports of infection in any mammalian host, using the virus name and relevant synonyms (www.ictvdb.org) as keywords in Web of Knowledge (http://wokinfo.com/), Wildlife Disease Association meeting abstracts (http://wildlifedisease.org/wda/CONFERENCES.aspx), Google Scholar (http://scholar.google.com/), and the Global Mammal Parasites Database (www.mammalparasites.org). The resulting 605 host–pathogen relationships included 56 unique viruses classified in 17 taxonomic families and 325 unique mammals classified in 15 taxonomic orders. We excluded rabies from our analysis because the intense research effort on this virus and its high pathogenicity in almost all of its wide range of hosts (7) would skew the data disproportionately.
We then conducted a secondary literature search to determine whether viruses in our database cause signs of disease in their wildlife hosts. For the search, we used an aggregate of all publications available in PubMed (www.ncbi.nlm.nih.gov/pubmed/), Web of Science (http://thomsonreuters.com/products_services/science/science_products/a-z/web_of_science/), BIOSIS Previews (http://thomsonreuters.com/products_services/science/science_products/a-z/biosis_previews/), and Biologic & Agricultural Index Plus (www.ebscohost.com/academic/biological-agricultural-index-plus); search terms consisted of virus names and International Committee on the Taxonomy of Viruses synonyms, host genus and species names, and common names [reconciled to the 2005 version of Mammal Species of the World (8)]. All resulting abstracts and available full text reports were examined until the first robust report of visible disease was encountered. Viruses were identified as causing visible disease in a host if individual or epizootic death or grossly visible or otherwise observable signs of illness (e.g., high fever, loss of mobility, or severe decline in body condition) were reported. A report was considered robust only if infections were confirmed by PCR analysis or virus isolation and clinical signs were explicitly recorded to have occurred during active infection. We excluded studies reporting only serologic findings because of potential cross-reactivity among related viruses and poor correlation between serologic status and concurrent infection. Our criteria of stopping a search once any evidence for visible disease was found meant that for mammal–virus pairs without visible disease, the search was exhaustive.
We considered diseases to be nonpathogenic in their hosts only if actively infected animals were explicitly reported to be free of visible disease. Animals with less clear signs of disease, such as nasal discharge or death of neonates, were not considered asymptomatic because of the low detection probability associated with these traits in wild mammal surveillance. We rejected reports of experimentally induced disease because of the risk that dosage and inoculation technique would not be consistent with naturally occurring infections. However, we included experimental studies if actively infected animals remained asymptomatic, with the assumption that 1) clinical signs of infection were most likely to be seen in animals monitored in laboratory settings than in the wild and 2) stressful conditions in captivity would heighten the likelihood of a normally benign pathogen leading to clinical signs (9). Furthermore, compared with naturally occurring infections, experimental infections often involve more direct routes of inoculation and are therefore more likely to induce disease.
We conducted a logistic regression analysis, using Firth’s bias reduction procedure (10) as used by the brglm (bias reduction in generalized linear models) package of R v2.15-2 (http://cran.r-project.org/bin/windows/base/), of apparent host disease as a function of host taxonomy and virus taxonomy for the subset of mammal–virus pairs for which the host order or virus family had at least 3 records in the database. We then calculated odds ratios for each host taxonomic order and virus family relative to the reference categories (Artiodactyla and Flaviviridae) and the predicted probability of being symptomatic for all species order–virus family combinations.
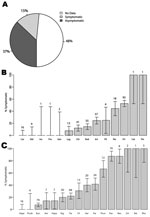
Our search of the 605 mammal–virus associations investigated yielded explicit information on host health in 52% of the 312 mammal–virus pairs. Of these, ≈28% (n = 88) of infected wildlife hosts were reported to have had visible disease and 72% (n = 224) were reported without evidence of visible disease (Figure 1, panel A). The proportion of hosts that were symptomatic differed across host order (Figure 1, panel B) and virus family (Figure 1, panel C).
We found that virus family and host order were significant predictors of disease status (χ2 = 88.70, p<0.001 and χ2 = 59.45, p<0.001, respectively). Species infected with paramyxoviruses, poxviruses, and reoviruses were more likely to have visible disease (p = 0.02, p = 0.001, and p = 0.04, respectively), and species infected with bunyaviruses were less likely to have visible disease relative to the reference category (p = 0.01) (Table). Hosts infected with filoviruses were marginally more likely to have visible disease (p = 0.08) (Table).
Relative to the reference category, species classified in the order Chiroptera were less likely to have visible disease (p<0.001), and species in the order Rodentia were marginally less likely to have visible disease (p = 0.10) (Table). Compared with species in other orders, species in the order Chiroptera had a lower probability of visible disease (Figure 2), although all Chiroptera species infected with nonrabies rhabdoviruses had a high probability of visible disease. In the dataset, all host–pairs infected with rhabdoviruses were in the order Chiroptera and were reported with visible disease in that host (Figure 1).
Nonhuman primates (11–13) and species classified within the taxonomic orders Chiroptera and Rodentia are the primary mammals targeted for zoonotic disease surveillance. Our data suggest that species in the orders Chiroptera and Rodentia are less likely than species in other orders to have visible disease (Figure 1). The mechanism behind this relationship is a critical area for additional research. In general, we found that the probability of having visible disease depends on the taxonomic classification of the host and virus, and Chiroptera is the only host order for which a single strategy (in this case, healthy animal surveillance) can be applied across nearly all virus families, excluding Rhabdoviridae. Therefore, particularly for the case of novel virus detection, our results point to a mixed strategy of targeted syndromic and healthy animal surveillance across host and virus taxonomies. A mixed strategy could combine apparently healthy animal surveillance (particularly in Chiroptera) with syndromic surveillance in other wildlife and domestic animal hosts. Syndromic surveillance has proven useful where secondary animal hosts are involved [e.g., surveillance for West Nile virus (14), henipaviruses (15,16), and Ebola virus (17)].
There are limitations to our study, particularly ascertainment and reporting biases, as acknowledged in previous studies of emerging infectious diseases (2,3). In addition, differences in the number of species belonging to each order, the difficulty of testing inaccessible species, and limits to reliable diagnoses of emerging viruses have an effect, especially in resource-poor settings. Furthermore, many disease states are not recognizable in free-ranging mammalian species under field conditions. Last, there is a risk that an animal may be co-infected with several agents, only one of which causes disease; that co-infection may have an additive or synergistic effect on clinical signs; and that anthropozoonotic viruses artificially inflate the disease count of mammals in some taxonomic orders over others. However, our findings were determined on the basis of an aggregation of the best data available on host health as it relates to zoonotic viruses, and they have useful implications for public health.
Our analysis supports a holistic, probability-based approach to zoonotic virus discovery, specifically, continued analysis of passively and actively reported deaths and increased investment in broad surveillance of healthy wildlife. The latter could be targeted geographically to those regions most likely to generate novel emerging infectious diseases (2) or taxonomically to groups that are reservoirs for the highest proportion of zoonoses (3,18). These efforts could be envisaged as part of a strategy for smart surveillance, heightening the opportunity for discovery of novel zoonoses, particularly if wildlife are sampled at key interfaces where contact with human or domestic animals (and thus the opportunity for spillover) is highest.
Ms Levinson recently completed her MSc in International Community Health at New York University, conducting research on patterns in emerging infectious viruses as a biological research associate at EcoHealth Alliance. She is currently an Urban Agriculture Volunteer in Kolda, Senegal, with Peace Corps Senegal and is interested in international health and development.
Dr Bogich is a disease ecologist and a Fogarty International RAPIDD postdoctoral research fellow in the Ecology and Evolutionary Biology Department, Princeton University and a scientific consultant for EcoHealth Alliance. Her primary research interests include the population dynamics of emerging infections and the optimal design of monitoring and management strategies for infectious disease control.
Acknowledgments
We acknowledge 2 anonymous reviewers and A. M. Kilpatrick for comments on the manuscript.
This study was funded in part by the generous support of the American people through the United States Agency for International Development (USAID) Emerging Pandemic Threats PREDICT program; a NIAID non-biodefense emerging infectious disease research opportunities award R01 AI079231 (to P.D.); the Research and Policy for Infectious Disease Dynamics (RAPIDD) program of the Science and Technology Directorate; the US Department of Homeland Security; the Fogarty International Center, National Institutes of Health (NIH) (to T.L.B.); and an NIH Fogarty American Recovery and Reinvestment Act award (3R01TW005869-06S10; to K.J.O).
References
- Taylor LH, Latham SM, Woolhouse MEJ. Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci. 2001;356:983–9. DOIPubMedGoogle Scholar
- Jones KE, Patel N, Levy M, Storeygard A, Balk D, Gittleman JL, Global trends in emerging infectious diseases. Nature. 2008;451:990–3. DOIPubMedGoogle Scholar
- Woolhouse MEJ, Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerg Infect Dis. 2005;11:1842–7 . DOIPubMedGoogle Scholar
- United States Agency for International Development. USAID launches Emerging Pandemic Threats program. 2009 [cited 2012 Jul 2]. http://www1.usaid.gov/press/releases/2009/pr091021_1.html.
- Morse SS. Emerging viruses. New York: Oxford University Press; 1993.
- Cleaveland S, Laurenson MK, Taylor LH. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Philos Trans R Soc Lond B Biol Sci. 2001;356:991–9. DOIPubMedGoogle Scholar
- Krebs JW, Wilson ML, Childs JI. Rabies: epidemiology, prevention, and future research. J Mammal. 1995;76:681–94 .DOIGoogle Scholar
- Wilson DE, Reeder DM, editors. Mammal species of the world. 3rd ed. Baltimore: Johns Hopkins University Press; 2005.
- Williams ES, Barker IK, editors. Infectious diseases of wild mammals. 3rd ed. Ames (IA): Iowa State Press; 2001.
- Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80:27–38. DOIGoogle Scholar
- Leendertz FH, Pauli G, Maetz-Rensing K, Boardman W, Nunn C, Ellerbrok H, Pathogens as drivers of population declines: the importance of systematic monitoring in great apes and other threatened mammals. Biol Conserv. 2006;131:325–37. DOIGoogle Scholar
- Wolfe ND, Daszak P, Kilpatrick AM, Burke DS. Bushmeat hunting, deforestation and prediction of zoonotic emergence. Emerg Infect Dis. 2005;11:1822–7. DOIPubMedGoogle Scholar
- Wolfe ND, Escalante AA, Karesh WB, Kilbourn A, Spielman A, Lal AA. Wild primate populations in emerging infectious disease research: the missing link? Emerg Infect Dis. 1998;4:149–58. DOIPubMedGoogle Scholar
- Eidson M, Komar N, Sorhage F, Nelson R, Talbot T, Mostashari F, Crow deaths as a sentinel surveillance system for West Nile virus in the northeastern United States, 1999. Emerg Infect Dis. 2001;7:615–20 .DOIPubMedGoogle Scholar
- Nor MNM, Gan CH, Ong BL. Nipah virus infection of pigs in peninsular Malaysia. Rev Sci Tech. 2000;19:160–5.PubMedGoogle Scholar
- Selvey LA, Wells RM, McCormack JG, Ansford AJ, Murray PK, Rogers RJ, Infection of humans and horses by a newly described morbillivirus. Med J Aust. 1995;162:642–5 .PubMedGoogle Scholar
- Leroy EM, Rouquet P, Formenty P, Souquière S, Kilbourne A, Froment JM, Multiple Ebola virus transmission events and rapid decline of central African wildlife. Science. 2004;303:387–90 . DOIPubMedGoogle Scholar
- Cooper N, Griffin R, Franz M, Omotayo M, Nunn CL. Phylogenetic host specificity and understanding parasite sharing in primates. Ecol Lett. 2012;15:1370–7. DOIPubMedGoogle Scholar
Figures
Table
Cite This Article1These authors contributed equally to this article.
Table of Contents – Volume 19, Number 5—May 2013
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Peter Daszak, EcoHealth Alliance, 460 W 34th St, 17th Flr, New York, NY 10001, USA
Top