Volume 20, Number 1—January 2014
Dispatch
Novel Avian Coronavirus and Fulminating Disease in Guinea Fowl, France
Figure 2
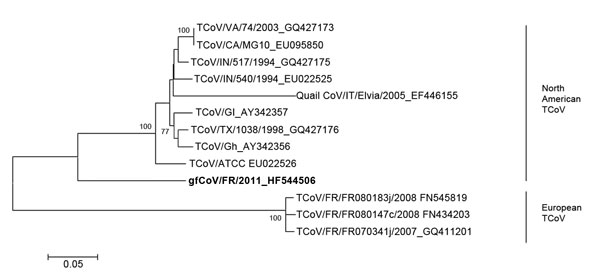
Figure 2. . . Phylogenetic analysis of the complete spike (S) gene of the fulminating disease of guinea fowl coronavirus (gfCoV, in boldface) in relation to turkey coronaviruses (TCoVs) at the nucleotide level. The tree was generated by using MEGA 5.05 (http://megasoftware.net/mega.php) by the neighbor-joining method, Kimura 2-parameter model, and pairwise deletion. Bootstrap values (1,000 replicates) >75 are indicated on the nodes. Only a partial S gene sequence (1,657 nt) was available for quail CoV/Italy/Elvia/2005. Scale bar indicates kimura distance.
1These authors contributed equally to this article.
Page created: January 03, 2014
Page updated: January 03, 2014
Page reviewed: January 03, 2014
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.