Volume 20, Number 3—March 2014
Dispatch
Influenza A(H1N1)pdm09 Virus Infection in Giant Pandas, China
Cite This Article
Citation for Media
Abstract
We confirmed infection with influenza A(H1N1)pdm09 in giant pandas in China during 2009 by using virus isolation and serologic analysis methods. This finding extends the host range of influenza viruses and indicates a need for increased surveillance for and control of influenza viruses among giant pandas.
In April 2009, the Centers for Disease Control and Prevention reported the emergence of a novel strain of influenza A(H1N1) virus, which is now referred to as influenza A(H1N1)pdm09 or pH1N1. This virus rapidly affected countries worldwide and continues to circulate as a seasonal influenza virus. In addition to widespread infection of humans, reported have been published of pH1N1 virus infection in domestic and nondomestic animals, including cats, dogs, ferrets, swine, and several wildlife species (1–4). Here, we report a confirmed case of pH1N1 virus infection in giant pandas (Ailuropoda melanoleuca) in China.
During the human outbreak of pH1N1 in China, 3 giant pandas at the Conservation and Research Center for the Giant Panda in Ya’an City, Sichuan Province, showed clinical signs suggestive of a respiratory tract infection, including pyrexia, anorexia, malaise, conjunctivitis, and sneezing. Nasal swab specimens were successfully collected under anesthesia from 1 of the affected pandas (Ximeng). All 3 pandas recovered after receiving 75 mg of oseltamivir phosphate twice daily for 5–6 days. For serologic analysis, serum samples were collected from all 3 pandas ≈3 months after the resolution of the respiratory illness; a serum sample collected before onset of the respiratory illness was also available for all 3 animals.
The nasal swab specimens were collected in 1 mL phosphate-buffered saline and tested for evidence of pH1N1 virus and several other pathogens reported (5–7) or suspected to occur in giant pandas: canine distemper virus, canine adenovirus, canine coronavirus, canine herpesvirus, and canine parainfluenza virus. Testing for detection of influenza A virus was performed by using a real-time reverse transcription PCR method, as described by the World Health Organization (WHO) (8); other pathogens were tested by different PCR methods.
RNA from the swab specimens tested positive for the hemagglutinin gene of pH1N1 virus. No other pathogens were detected.
To isolate and characterize the pH1N1 virus, we injected 10-day-old specific pathogen free embryonated chicken eggs with material collected from one of the nasal swab samples. Allantoic fluid from the injected eggs agglutinated 0.5% (vol/vol) chicken erythrocytes, indicating the presence of replication-competent virus in the nasal swab sample. When evaluated by electron microscopy, allantoic fluid supernatant from the infected egg displayed enveloped influenza virus–like particles of 100–120 nm (Figure 1).
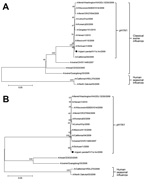
We sequenced the entire genome of the virus by using RNA harvested from the allantoic fluid of infected eggs. Sequence analysis was performed as described by WHO by using methods for pH1N1 virus (9). A BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to identify sequences similar to those of the giant panda isolate; these sequences were GenBank accession nos. KF277197–KF277204. Analyses showed that each of the 8 gene segments of the virus we isolated were closely related to pH1N1 viruses circulating among humans, including a human representative strain (A/California/04/2009) and a contemporary strain (A/Sichuan/1/2009); these viruses showed 98.6%–100% nt identity to the panda strain (Technical Appendix Table 1). Phylogenetic analysis of the 8 gene segments of the virus we isolated showed that the isolate was related to the pH1N1 virus clade of influenza viruses (Figure 2; Technical Appendix Figure). The results of the BLAST and phylogenetic analyses suggest that a human pH1N1 virus was transmitted directly to giant pandas without recombination or significant adaptation. We named the virus that we isolated A/giant panda/01/Ya’an/2009 (H1N1).
A/giant panda/01/Ya’an/2009 (H1N1) virus contained glutamine at hemagglutinin amino acid position 226 (H3 numbering), which has been found in human pH1N1 isolates and confers binding to human cell-surface receptors (10). In addition, alanine at position 271 of polymerase protein 2 of the isolate reported here has been shown to be required for respiratory droplet–mediated transmission of A/Sichuan/01/2009 virus (10). Thus, the genomic sequences of the giant panda isolate appear to have many features known to be important for the replication and transmission of pH1N1 viruses in other mammalian species.
Serum samples collected from the affected pandas before their illnesses and 3 months after the resolution of the illnesses were tested for the presence of hemagglutination inhibition (HI) antibodies against a panel of influenza A viruses. HI antibody titers were measured against the A/giant panda/01/Ya’an/2009 (H1N1) isolate, human seasonal influenza (H3N2) virus (A/Victoria/361/2011), avian (H5N1) virus (A/duck/Anhui/1/2006), avian (H6N1) virus (A/Mallard/SanJiang/275/2007), and avian (H7N1) virus (A/Baer’s pochard/HuNan/414/2010). All 3 pandas had high titers of HI antibodies against the pH1N1 virus 3 months after illness (Table).
One panda (Zhangka) had detectable pH1N1 and H3 subtype HI antibodies before infection; this serum sample was collected before the display of overt clinical signs in the animal but after the onset of the human pH1N1 outbreak in China. It is unknown if these antibodies reflect previous exposure to pH1N1 virus or are reflective of cross-reactive antibodies generated against antigenic sites in previously circulating influenza viruses.
Two of the pandas (Gege and Zhangka) also had serum antibodies that inhibited hemagglutination mediated by an H6 subtype avian influenza virus 3 months after the respiratory infection, whereas no such antibodies were detected before the respiratory infection (Table). When reference serum samples known to contain HI antibodies against each of the viral subtypes were evaluated for potential cross-reactivity against the other influenza subtypes, we observed no apparent cross-reactivity of pH1N1 antibodies against the H6 subtype virus (Technical Appendix Table 2). Therefore, although it is possible that the high titers of HI antibodies against pH1N1 virus in these animals cross-reacted with the H6 subtype viruses, we cannot exclude the possibility that these antibodies were generated in response to an independent exposure to an H6 influenza virus.
Influenza A(H1N1)pdm09 virus infection has been found in mammals and birds of a variety of species since the virus was identified in early 2009 (1–4). We documented infection with this virus in giant pandas in China during 2009. Sequence analysis of a virus isolated from a nasal swab sample revealed that it was nearly identical to the pH1N1 virus circulating in humans, which suggests that the virus may have been transmitted to the pandas directly from humans. Furthermore, serologic analysis of samples from the affected pandas was consistent with productive infection by pH1N1 virus. Seroconversion was documented in each animal, although 1 animal appeared to have been previously exposed to pH1N1 or a related virus that was able to elicit cross-reactive antibodies. Similarly, some pandas had detectable HI antibodies against H3 and H6 subtype influenza viruses, raising the possibility that giant pandas may be infected with human- and avian-origin influenza viruses.
Only 1,600 wild and 300 captive giant pandas remain worldwide (11). Our findings highlight the risk posed by influenza viruses to these animals, extend the known host range of influenza A viruses, and have implications for wildlife conservation and influenza virus epidemiology. When taken together with previous reports of pH1N1 infection of other mammals, our data suggest that this virus can efficiently infect a wider range of mammalian species than can other influenza viruses. Increased surveillance and control of influenza viruses in giant panda populations are imperative for successful conservation of these endangered animals.
Dr Li is deputy director at the China Conservation and Research Center for the Giant Panda, Ya’An, People’s Republic of China. His main research interest is conservation of giant pandas.
Acknowledgments
We thank Peter Wilker for editing the manuscript.
This work was supported by the State Forestry Administration international collaboration project (no. WH1110), Wolong Giant Panda Club Foundation (no. 201001), 973 Program (no. 2011CB50502), and 863 program (no. 2012AA022006).
References
- Howden KJ, Brockhoff EJ, Caya FD, McLeod LJ, Lavoie M, Ing JD, An investigation into human pandemic influenza virus (H1N1) 2009 on an Alberta swine farm. Can Vet J. 2009;50:1153–61.PubMedGoogle Scholar
- Sponseller BA, Strait E, Jergens A, Trujillo J, Harmon K, Koster L, Influenza A pandemic (H1N1) 2009 virus infection in domestic cat. Emerg Infect Dis. 2010;16:534–7. DOIPubMedGoogle Scholar
- Lin D, Sun S, Du L, Ma J, Fan L, Pu J, Natural and experimental infection of dogs with pandemic H1N1/2009 influenza virus. J Gen Virol. 2012;93:119–23. DOIPubMedGoogle Scholar
- Britton AP, Sojonky KR, Scouras AP, Bidulka JJ. Pandemic (H1N1) 2009 in skunks, Canada. Emerg Infect Dis. 2010;16:1043–5. DOIPubMedGoogle Scholar
- Loeffler IK, Howard J, Montali RJ, Hayek LA, Dubovi E, Zhang Z, Serosurvey of ex situ giant pandas (Ailuropoda melanoleuca) and red pandas (Ailurus fulgens) in China with implications for species conservation. J Zoo Wildl Med. 2007;38:559–66. DOIPubMedGoogle Scholar
- Gao FS, Hu GX, Xia XZ, Gao YW, Bai YD, Zou XH. Isolation and identification of a canine coronavirus strain from giant pandas (Ailuropoda melanoleuca). J Vet Sci. 2009;10:261–3. DOIPubMedGoogle Scholar
- Qin Q, Li D, Zhang H, Hou R, Zhang Z, Zhang C, Serosurvey of selected viruses in captive giant pandas (Ailuropoda melanoleuca) in China. Vet Microbiol. 2010;142:199–204. DOIPubMedGoogle Scholar
- World Health Organization. CDC protocol of realtime RTPCR for influenza A(H1N1). 2009 Oct 6 [cited 2009 Dec 15]. http://www.who.int/csr/resources/publications/swineflu/CDCRealtimeRTPCR_SwineH1Assay-2009_20090430.pdf
- World Health Organization. Sequencing primers and protocol. 2009 May 12 [cited 2009 Dec 15]. http://www.who.int/csr/resources/publications/swineflu/GenomePrimers_20090512.pdf
- Zhang Y, Zhang Q, Gao Y, He X, Kong H, Jiang Y, Key molecular factors in hemagglutinin and PB2 contribute to efficient transmission of the 2009 H1N1 pandemic influenza virus. J Virol. 2012;86:9666–74. DOIPubMedGoogle Scholar
- Lai F, Olesen B. A visual celebration of giant pandas. Singapore: Editions Didier Millet; 2013. p. 151.
Figures
Table
Cite This Article1These authors contributed equally to this article.
Table of Contents – Volume 20, Number 3—March 2014
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Yuwei Gao, Research Center of Wildlife Disease, Key Laboratory of Jilin Province for Zoonosis Prevention and Control, Military Veterinary Research Institute, Academy of Military Medical Sciences, 666 Liuyingxi Rd, Changchun 130122, People’s Republic of ChinaYuwei Gao, Research Center of Wildlife Disease, Key Laboratory of Jilin Province for Zoonosis Prevention and Control, Military Veterinary Research Institute, Academy of Military Medical Sciences, 666 Liuyingxi Rd, Changchun 130122, People’s Republic of China
Top