Volume 20, Number 7—July 2014
Dispatch
Detection and Genetic Characterization of Deltacoronavirus in Pigs, Ohio, USA, 2014
Figure 1
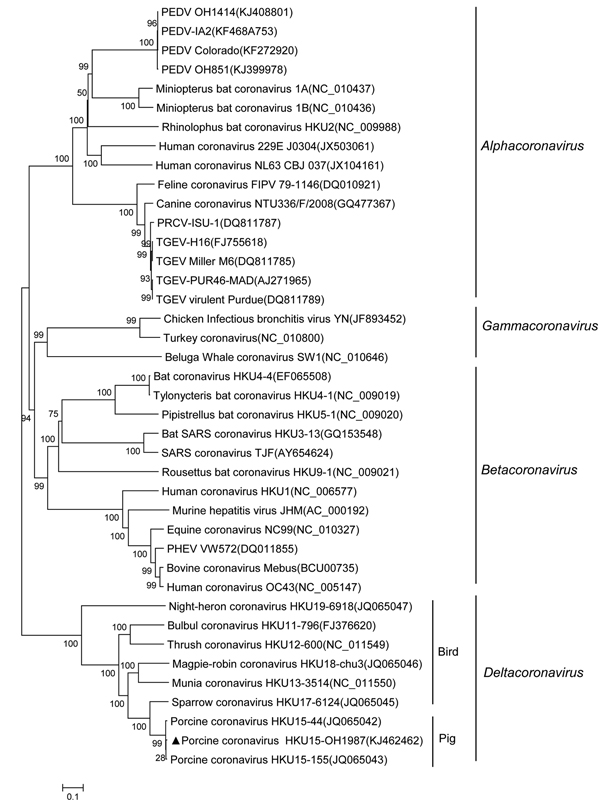
Figure 1. Phylogenetic tree constructed on the basis of the whole-genome sequences of virus strains from 4 coronavirus genera (Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus), including the porcine coronavirus HKU15 OH1987 strain (indicated with triangle)The dendrogram was constructed by using the neighbor-joining method in the MEGA software package, version 6.05 (http://www.megasoftware.net/)Bootstrap resampling (1,000 replications) was performed, and bootstrap values are indicated for each nodeReference sequences obtained from GenBank are indicated by strain name and accession numberScale bar represents 0.1 nt substitutions per sitePEDV, virus and porcine epidemic diarrhea virus; PRCV, porcine respiratory coronavirus; TGEV, transmissible gastroenteritis virus; SARS, severe acute respiratory syndromePHEV, porcine hemagglutinating encephalomyelitis virus.
1These authors were co–principal investigators.