Volume 20, Number 8—August 2014
Research
Rapid Whole-Genome Sequencing for Surveillance of Salmonella enterica Serovar Enteritidis
Figure 1
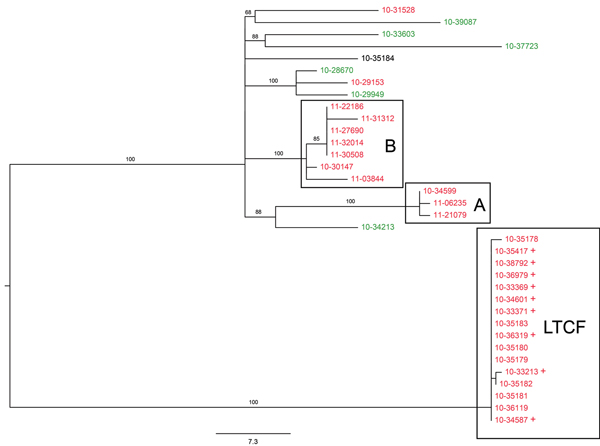
Figure 1. Figure 1. Maximum-likelihood tree of population structure of Salmonella enterica serovar Enteritidis isolates obtained in New York and neighboring states, USA. The tree was inferred by using a general time-reversible model with a gamma distribution and was inferred to be the best fit model by the maximum-likelihood method implemented in MEGA 5.1 (22). Values on branches are bootstrap values based on 150 bootstrap replicates. Note the well supported and distant cluster associated with the long-term care facility (LTCF), as well as additional clusters A and B. Labels of isolates are colored according to their New York State Department of Health Wadsworth Laboratories multilocus variable-number tandem-repeat analysis (MLVA) subtype designation. Green, MLVA subtype B; red, MLVA subtype W; black, MLVA subtype AE. + indicates isolates in the LTCF cluster that were detected only by whole-genome analysis and were not detected epidemiologically. Scale bar indicates single-nucleotide polymorphisms per site.