Volume 20, Number 8—August 2014
Dispatch
Levofloxacin-Resistant Haemophilus influenzae, Taiwan, 2004–2010
Figure
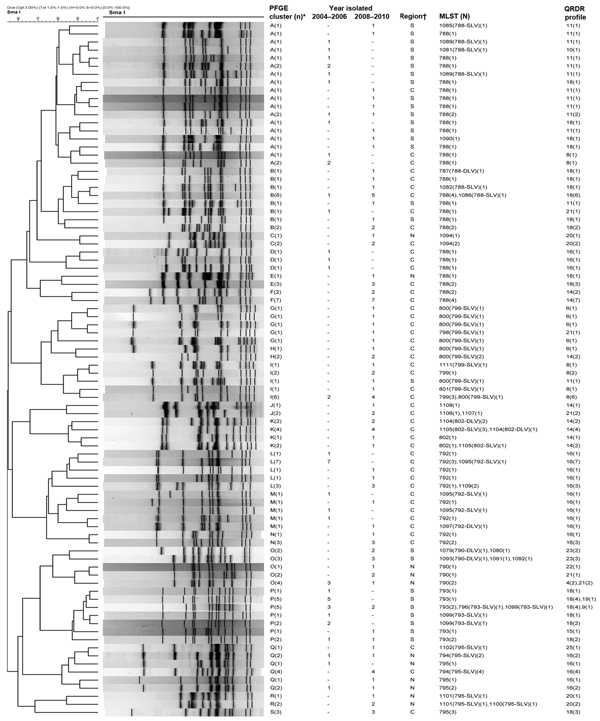
Figure. Dendrogram showing pulsed-field gel electrophoresis (PFGE) results for levofloxacin-resistant Haemophilus influenzae isolates digested by SmaISalmonella enterica serovar Braenderup strain H9812 (ATCC BAA664) was used as standard for DNA pattern normalizationPFGE patterns were analyzed by using BioNumerics software (Applied Maths NV, Sint-Martens-Latem, Belgium)For mutation profiles of the quinolone-resistance determining regions (QRDR) in GyrA and ParC, see Table 3*Isolates having >80% similarity or <6 band differences were assigned a PFGE cluster if there were >3 isolates within the cluster; †region of hospital location: C, central; N, north; S, south–, no isolates found(n), number of isolates having the same PFGE pattern; MLST, multilocus sequence typing; (N), number of isolates on which MLST was performed; SLV, single locus variant, DLV, double locus variant.