Volume 20, Number 9—September 2014
Research
Genomic Epidemiology of Salmonella enterica Serotype Enteritidis based on Population Structure of Prevalent Lineages
Figure 2
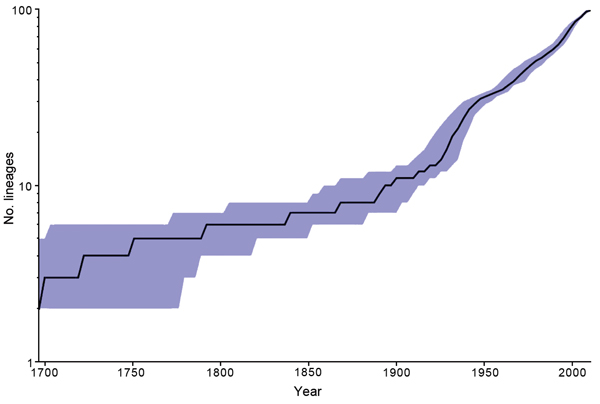
Figure 2. Inferred number of Salmonella enterica serotype Enteritidis lineages over time based on a constant effective population size model using BEAST (16). Blue shading indicates 95% CIs.
1These authors contributed equally to this article.
Page created: August 13, 2014
Page updated: August 13, 2014
Page reviewed: August 13, 2014
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.