Volume 21, Number 12—December 2015
Dispatch
Water as Source of Francisella tularensis Infection in Humans, Turkey
Figure 1
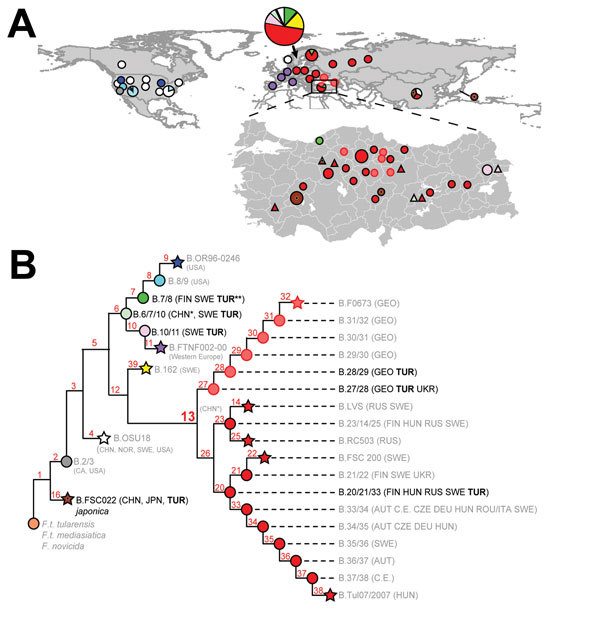
Figure 1. Phylogeography of Francisella tularensis subsp. holarctica. A) Global distribution of known phylogenetic groups determined on the basis of previous studies (2–4); enlarged map of Turkey shows locations of phylogenetic groups identified among the 40 samples positive for F. tularensis examined in this and previous studies (5). Circle size indicates number of samples (small circles, 1–3; medium circles, 4–6; large circles, 7–9). Colors of circles (human samples) and triangles (environmental samples) represent the phylogenetic subgroups to which these samples were assigned (see panel B). Subgroup B.16 (biovar japonica) is represented by the dot inside the brown circles and triangles. B) Phylogenetic tree for F. tularensis subsp. holarctica constructed on the basis of current canonical single-nucleotide polymorphism genotyping. Red numbers indicate nomenclature of canonical single-nucleotide polymorphism groups. Terminal subgroups representing sequenced strains are shown as stars, and intervening nodes representing collapsed branches are indicated by circles. Countries of origin for samples assigned to relevant phylogenetic groups are as follows: AUT, Austria; CE, central Europe, unknown country; CHN, China; CZE, Czech Republic; DEU, Germany; FIN, Finland; GEO, Georgia; HUN, Hungary; ITA, Italy; NOR Norway; ROU, Romania; RUS, Russia; SWE, Sweden; TUR, Turkey; UKR, Ukraine; USA, United States. CHN* indicates approximate phylogenetic placement because of a lack of resolved information on single-nucleotide polymorphisms (4). TUR** indicates identification from a previous study (5).
References
- Vogler AJ, Birdsell D, Price LB, Bowers JR, Beckstrom-Sternberg SM, Auerbach RK, Phylogeography of Francisella tularensis: global expansion of a highly fit clone. J Bacteriol. 2009;191:2474–84. DOIPubMedGoogle Scholar
- Karlsson E, Svensson K, Lindgren P, Byström M, Sjödin A, Forsman M, The phylogeographic pattern of Francisella tularensis in Sweden indicates a Scandinavian origin of Eurosiberian tularaemia. Environ Microbiol. 2013;15:634–45. DOIPubMedGoogle Scholar
- Gyuranecz M, Birdsell DN, Splettstoesser W, Seibold E, Beckstrom-Sternberg SM, Makrai L, Phylogeography of Francisella tularensis subsp. holarctica, Europe. Emerg Infect Dis. 2012;18:290–3. DOIPubMedGoogle Scholar
- Wang Y, Peng Y, Hai R, Xia L, Li H, Zhang Z, Diversity of Francisella tularensis subsp. holarctica lineages, China. Emerg Infect Dis. 2014;20:1191–4 . DOIPubMedGoogle Scholar
- Özsürekci Y, Birdsell DN, Çelik M, Karadağ-Öncel E, Johansson A, Forsman M, Phylogenetically diverse Francisella tularensis strains cause human tularemia in Turkey. Emerg Infect Dis. 2015;21:173–5. DOIPubMedGoogle Scholar
- Kiliç S, Celebi B, Acar B, Atas M. In vitro susceptibility of isolates of Francisella tularensis from Turkey. Scand J Infect Dis. 2013;45:337–41. DOIPubMedGoogle Scholar
- Kiliç S. Tularemia: the pathogen and epidemiology [in Turkish]. Turkiye Klinikleri J.E.N.T–Special Topics. 2014;7:52–61.
- Erdem H, Ozturk-Engin D, Yesilyurt M, Karabay O, Elaldi N, Celebi G, Evaluation of tularaemia courses: a multicentre study from Turkey. Clin Microbiol Infect. 2014;20:O1042–51. DOIPubMedGoogle Scholar
- Willke A, Meric M, Grunow R, Sayan M, Finke EJ, Splettstößer W, An outbreak of oropharyngeal tularaemia linked to natural spring water. J Med Microbiol. 2009;58:112–6. DOIPubMedGoogle Scholar
- Ulu Kiliç A, Kiliç S, Sencan I, Cicek Sentürk G, Gürbüz Y, Tütüncü EE, A water-borne tularemia outbreak caused by Francisella tularensis subspecies holarctica in Central Anatolia region [in Turkish]. Mikrobiyol Bul. 2011;45:234–47.PubMedGoogle Scholar
- Unal Yilmaz G, Gurcan S, Ozkan B, Karadenizli A. Investigation of the presence of Francisella tularensis by culture, serology and molecular methods in mice of Thrace Region, Turkey [in Turkish]. Mikrobiyol Bul. 2014;48:213–22. DOIPubMedGoogle Scholar
- Sjöstedt A, Kuoppa K, Johansson T, Sandströom G. The 17 kDa lipoprotein and encoding gene of Francisella tularensis LVS are conserved in strains of Francisella tularensis. Microb PathogDOIPubMedGoogle Scholar
- Vogler AJ, Birdsell D, Wagner DM, Keim P. An optimized, multiplexed multi-locus variable-number tandem repeat analysis system for genotyping Francisella tularensis. Lett Appl Microbiol. 2009;48:140–4. DOIPubMedGoogle Scholar
- Christova I, Gladnishka T. Prevalence of infection with Francisella tularensis, Borrelia burgdorferi sensu lato and Anaplasma phagocytophilum in rodents from an endemic focus of tularemia in Bulgaria. Ann Agric Environ Med. 2005;12:149–52.PubMedGoogle Scholar
- Johansson A, Lärkeryd A, Widerström M, Mörtberg S, Myrtännäs K, Ohrman C, An outbreak of respiratory tularemia caused by diverse clones of Francisella tularensis. Clin Infect Dis. 2014;59:1546–53. DOIPubMedGoogle Scholar
1These authors contributed equally to this article.