Volume 21, Number 12—December 2015
Dispatch
Water as Source of Francisella tularensis Infection in Humans, Turkey
Figure 2
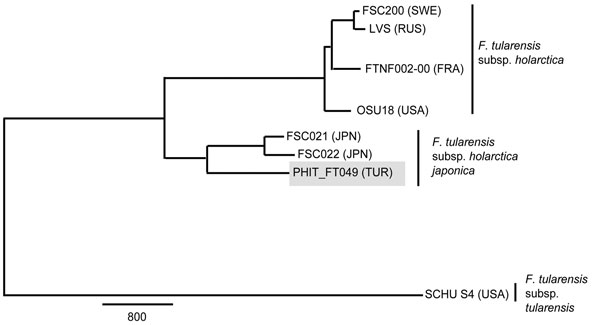
Figure 2. Maximum-parsimony phylogeny constructed by using 10,443 putative single-nucleotide polymorphisms discovered from whole-genome sequences of 8 Francisella tularensis strains. Gray shading indicates the B.16 (biovar japonica) strain from Turkey (PHIT_FT049). Detailed methods are described in the Technical Appendix. Reference strains were retrieved from GenBank (Technical Appendix Table 2). Countries of origin are indicated as follows: FRA, France; JPN, Japan; RUS, Russia; SWE, Sweden; TUR, Turkey; USA, United States. Scale bar indicates single-nucleotide polymorphisms.
1These authors contributed equally to this article.
Page created: November 17, 2015
Page updated: November 17, 2015
Page reviewed: November 17, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.