Volume 21, Number 12—December 2015
Dispatch
Asymptomatic MERS-CoV Infection in Humans Possibly Linked to Infected Dromedaries Imported from Oman to United Arab Emirates, May 2015
Figure
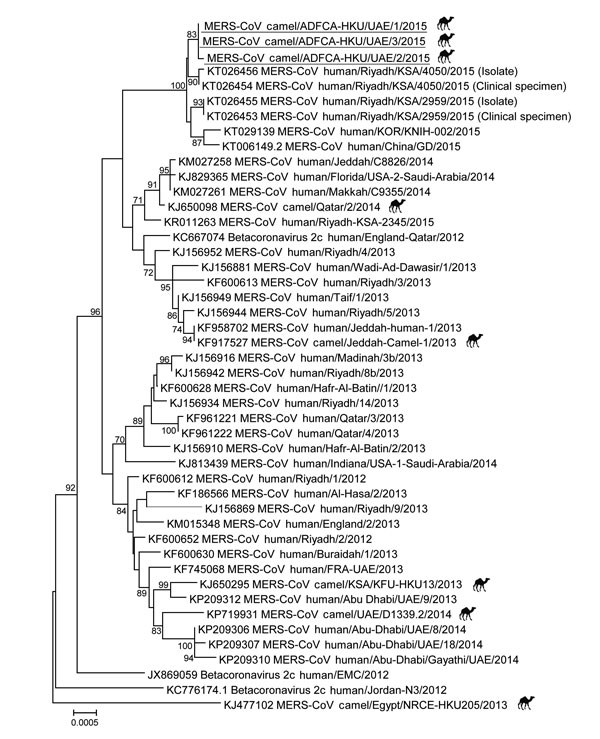
Figure. Phylogenetic analyses of partial Middle East respiratory syndrome coronavirus (MERS-CoV) genomic sequences for viruses detected in dromedary camels imported from Oman to United Arab Emirates, May 2015. A partial viral RNA sequence spanning the 3′ end of the open reading frame 1AB gene through the 3′ untranslated region of the MERS-CoV genome (≈8,900 nt) was used in the analysis. The phylogenetic tree was constructed with MEGA6 software (http://www.megasoftware.net/) by using the neighbor-joining method. Numbers at nodes indicate bootstrap values determined by 1,000 replicates. Only bootstrap values >70 are denoted. Underlining indicates sequences for viruses detected in this study. GenBank accession numbers are shown for published sequences. Camel symbols indicate MERS-CoVs detected from camels. Scale bar indicates the estimated genetic distance of these viruses.
1These first authors contributed equally to this article.