Volume 21, Number 2—February 2015
Research
Evidence for Elizabethkingia anophelis Transmission from Mother to Infant, Hong Kong
Figure 2
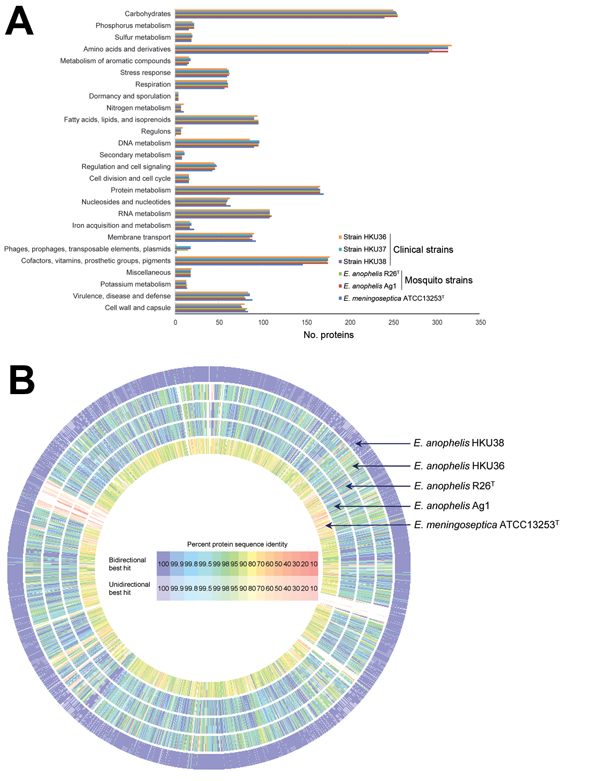
Figure 2. Comparison of draft genome sequence data of the 3 Elizabethkingia anophelis strains from patients in Hong Kong (HKU36–38), E anophelis type strain R26T, and E. meningoseptica type strain ATCC 13253T. A) Distributions of predicted coding sequence function in genomes of E. anophelis strains HKU36–38, E. anophelis type strain R26T, and E. meningoseptica type strain ATCC 13253T according to SEED Subsystems are shown. The columns indicate the number of proteins in different subsystems. B) Circular representation of sequence comparison between the draft genome of strain HKU37 and other draft genomes as labeled. Comparison generated in Rapid Annotations using Subsystem Technology (27). Intensity of color indicates degree of protein identity.
References
- Loman NJ, Constantinidou C, Chan JZ, Halachev M, Sergeant M, Penn CW, High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nat Rev Microbiol. 2012;10:599–606 . DOIPubMedGoogle Scholar
- Fournier PE, Drancourt M, Raoult D. Bacterial genome sequencing and its use in infectious diseases. Lancet Infect Dis. 2007;7:711–23 . DOIPubMedGoogle Scholar
- Shah MA, Mutreja A, Thomson N, Baker S, Parkhill J, Dougan G, Genomic epidemiology of Vibrio cholerae O1 associated with floods, Pakistan, 2010. Emerg Infect Dis. 2014;20:13–20 . DOIPubMedGoogle Scholar
- Snyder LA, Loman NJ, Faraj LA, Levi K, Weinstock G, Boswell TC, Epidemiological investigation of Pseudomonas aeruginosa isolates from a six-year-long hospital outbreak using high-throughput whole genome sequencing. Euro Surveill. 2013;18:20611 .PubMedGoogle Scholar
- Harris SR, Cartwright EJ, Török ME, Holden MT, Brown NM, Ogilvy-Stuart AL, Whole-genome sequencing for analysis of an outbreak of methicillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis. 2013;13:130–6 . DOIPubMedGoogle Scholar
- Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13:137–46. DOIPubMedGoogle Scholar
- Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet. 2013;381:1551–60. DOIPubMedGoogle Scholar
- Tse H, Bao JY, Davies MR, Maamary P, Tsoi HW, Tong AH, Molecular characterization of the 2011 Hong Kong scarlet fever outbreak. J Infect Dis. 2012;206:341–51. DOIPubMedGoogle Scholar
- Brown CC, Olsen RJ, Fittipaldi N, Morman ML, Fort PL, Neuwirth R, Spread of virulent group A Streptococcus type emm59 from Montana to Wyoming, USA. Emerg Infect Dis. 2014;20:679–81 . DOIPubMedGoogle Scholar
- Köser CU, Holden MT, Ellington MJ, Cartwright EJ, Brown NM, Ogilvy-Stuart AL, Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med. 2012;366:2267–75 . DOIPubMedGoogle Scholar
- Loman NJ, Constantinidou C, Christner M, Rohde H, Chan JZ, Quick J, A culture-independent sequence-based metagenomics approach to the investigation of an outbreak of Shiga-toxigenic Escherichia coli O104:H4. JAMA. 2013;309:1502–10. DOIPubMedGoogle Scholar
- Price JR, Golubchik T, Cole K, Wilson DJ, Crook DW, Thwaites GE, Whole-genome sequencing shows that patient-to-patient transmission rarely accounts for acquisition of Staphylococcus aureus in an intensive care unit. Clin Infect Dis. 2014;58:609–18. DOIPubMedGoogle Scholar
- Pérez-Lago L, Comas I, Navarro Y, González-Candelas F, Herranz M, Bouza E, Whole genome sequencing analysis of intrapatient microevolution in Mycobacterium tuberculosis: potential impact on the inference of tuberculosis transmission. J Infect Dis. 2014;209:98–108 . DOIPubMedGoogle Scholar
- Johnson PD, Ballard SA, Grabsch EA, Stinear TP, Seemann T, Young HL, A sustained hospital outbreak of vancomycin-resistant Enterococcus faecium bacteremia due to emergence of vanB E. faecium sequence type 203. J Infect Dis. 2010;202:1278–86 . DOIPubMedGoogle Scholar
- Kämpfer P, Matthews H, Glaeser SP, Martin K, Lodders N, Faye I. Elizabethkingia anophelis sp. nov., isolated from the midgut of the mosquito Anopheles gambiae. Int J Syst Evol Microbiol. 2011;61:2670–5. DOIPubMedGoogle Scholar
- Kim KK, Kim MK, Lim JH, Park HY, Lee ST. Transfer of Chryseobacterium meningosepticum and Chryseobacterium miricola to Elizabethkingia gen. nov. as Elizabethkingia meningoseptica comb. nov. and Elizabethkingia miricola comb. nov. Int J Syst Evol Microbiol. 2005;55:1287–93. DOIPubMedGoogle Scholar
- Frank T, Gody JC, Nguyen LB, Berthet N, Le Fleche-Mateos A, Bata P, First case of Elizabethkingia anophelis meningitis in the Central African Republic. Lancet. 2013;381:1876. DOIPubMedGoogle Scholar
- Bobossi-Serengbe G, Gody JC, Beyam NE, Bercion R. First documented case of Chryseobacterium meningosepticum meningitis in Central African Republic. Med Trop (Mars). 2006;66:182–4 .PubMedGoogle Scholar
- Teo J, Tan SY, Tay M, Ding Y, Kjelleberg S, Givskov M, First case of E anophelis outbreak in an intensive-care unit. Lancet. 2013;382:855–6. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial disk susceptibility tests. Approved standard, 11th ed. M02–A11. Wayne (PA): The Institute; 2012.
- Lau SK, Tang BS, Curreem SO, Chan TM, Martelli P, Tse CW, Matrix-assisted laser desorption ionization–time of flight mass spectrometry for rapid identification of Burkholderia pseudomallei: importance of expanding databases with pathogens endemic to different localities. J Clin Microbiol. 2012;50:3142–3. DOIPubMedGoogle Scholar
- Woo PC, Lau SK, Teng JL, Que TL, Yung RW, Luk WK, L Hongkongensis study group. Association of Laribacter hongkongensis in community-acquired gastroenteritis with travel and eating fish: a multicentre case-control study. Lancet. 2004;363:1941–7. DOIPubMedGoogle Scholar
- Lau SK, Curreem SO, Lin CC, Fung AM, Yuen KY, Woo PC. Streptococcus hongkongensis sp. nov., isolated from a patient with an infected puncture wound and from a marine flatfish. Int J Syst Evol Microbiol. 2013;63:2570–6. DOIPubMedGoogle Scholar
- Tse H, Tsoi HW, Leung SP, Lau SK, Woo PC, Yuen KY. Complete genome sequence of Staphylococcus lugdunensis strain HKU09–01. J Bacteriol. 2010;192:1471–2. DOIPubMedGoogle Scholar
- Woo PC, Lau SK, Tse H, Teng JL, Curreem SO, Tsang AK, The complete genome and proteome of Laribacter hongkongensis reveal potential mechanisms for adaptations to different temperatures and habitats. PLoS Genet. 2009;5:e1000416. DOIPubMedGoogle Scholar
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–9. DOIPubMedGoogle Scholar
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. DOIPubMedGoogle Scholar
- Liu B, Pop M. ARDB—antibiotic resistance genes database. Nucleic Acids Res. 2009;37:D443–7 . DOIPubMedGoogle Scholar
- Auch AF, Klenk HP, Göker M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci. 2010;2:142–8. DOIPubMedGoogle Scholar
- Dussurget O. New insights into determinants of Listeria monocytogenes virulence. Int Rev Cell Mol Biol. 2008;270:1–38.
- Kreft J, Vázquez-Boland JA, Altrock S, Dominguez-Bernal G, Goebel W. Pathogenicity islands and other virulence elements in Listeria. Curr Top Microbiol Immunol. 2002;264:109–25 .PubMedGoogle Scholar
- Hoffman JA, Badger JL, Zhang Y, Huang SH, Kim KS. Escherichia coli K1 aslA contributes to invasion of brain microvascular endothelial cells in vitro and in vivo. Infect Immun. 2000;68:5062–7. DOIPubMedGoogle Scholar
- Cheng C, Chen J, Fang C, Xia Y, Shan Y, Liu Y, Listeria monocytogenes aguA1, but not aguA2, encodes a functional agmatine deiminase: biochemical characterization of its catalytic properties and roles in acid tolerance. J Biol Chem. 2013;288:26606–15. DOIPubMedGoogle Scholar
- Matyi SA, Hoyt PR, Hosoyama A, Yamazoe A, Fujita N, Gustafson JE. Draft genome sequences of Elizabethkingia meningoseptica. Genome Announc. 2013;1:e00444–13.
- Kukutla P, Lindberg BG, Pei D, Rayl M, Yu W, Steritz M, Draft genome sequences of Elizabethkingia anophelis strains R26T and Ag1 from the midgut of the malaria mosquito Anopheles gambiae. Genome Announc. 2013;1:e01030–13.
- Balm MN, Salmon S, Jureen R, Teo C, Mahdi R, Seetoh T, Bad design, bad practices, bad bugs: frustrations in controlling an outbreak of Elizabethkingia meningoseptica in intensive care units. J Hosp Infect. 2013;85:134–40. DOIPubMedGoogle Scholar
- Quick J, Constantinidou C, Pallen MJ, Oppenheim B, Loman NJ. Draft genome sequence of Elizabethkingia meningoseptica isolated from a traumatic wound. Genome Announc. 2014;2:e00355–14.
- Sarma S, Kumar N, Jha A, Baveja U, Sharma S. Elizabethkingia meningosepticum: an emerging cause of septicemia in critically ill patients. J Lab Physicians. 2011;3:62–3. DOIPubMedGoogle Scholar
- Teo J, Tan SY, Liu Y, Tay M, Ding Y, Li Y, Comparative genomic analysis of malaria mosquito vector-associated novel pathogen Elizabethkingia anophelis. Genome Biol Evol. 2014;6:1158–65. DOIPubMedGoogle Scholar
- Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A. 2004;101:9786–91. DOIPubMedGoogle Scholar
1These authors contributed equally to this article.