Volume 21, Number 8—August 2015
Dispatch
Genomic Assays for Identification of Chikungunya Virus in Blood Donors, Puerto Rico, 2014
Figure 1
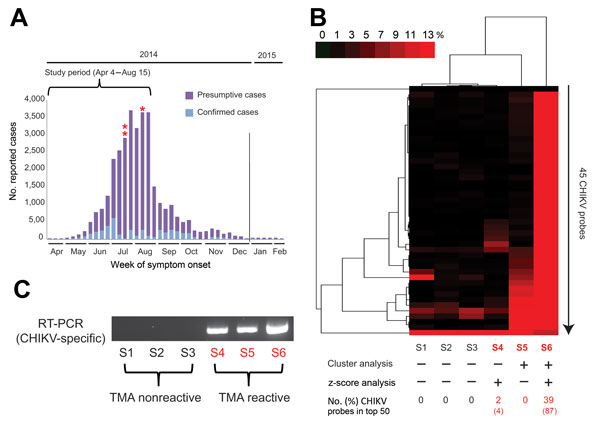
Figure 1. New genomic tests for chikungunya (CHIKV) infection in blood donors. A) Epidemic curve of reported cases in Puerto Rico, April 2014–February 2015. For 2014, 30,983 presumptive cases and 4,275 laboratory-confirmed cases were reported to the Secretary of Health in Puerto Rico. Three CHIKV-positive case-patients (asterisks) of 557 tested were identified by transcription-mediated amplification (TMA) screening of plasma samples during the study period. B) Heat map (cluster analysis) of 6 ViroChip (University of California San Francisco, San Francisco, CA, USA) microarrays corresponding to 6 donor plasma samples, 3 CHIKV positive and 3 CHIKV negative. Only microarray probes derived from CHIKV are plotted because signatures for other bloodborne viral pathogens were absent (data not shown). A sample is called ViroChip positive for CHIKV if at least 10% of the CHIKV probes on the heat map have a normalized probe intensity of >10% by cluster analysis (5) and/or if >1 probe is detected within the top 50 by z score analysis (6). Red bar denotes the magnitude of hybridization intensity normalized across the 45 CHIKV probes on the microarray. ViroChip microarray data have been submitted to the Gene Expression Omnibus database repository (accession no. GSE67234). C) Reverse transcription PCR (RT-PCR) testing for CHIKV and visualization of the PCR amplicon by 2% agarose gel electrophoresis confirm the transcription-mediated and ViroChip microarray results (7). D) Metagenomic next-generation sequencing (NGS) of the 3 CHIKV-positive plasma samples enables recovery of the viral genome. For each sample, coverage plots of mapped NGS reads to the “best hit” viral genome (accession no. KJ451624), identified by using the automated sequence-based ultrarapid pathogen identification pipeline, are shown (8). The read coverage (y axis, log scale) is plotted as a function of nucleotide position along the genome (x axis). The consensus whole-genome sequences obtained from the coverage plots are used for the subsequent phylogenetic and molecular clock analyses (Figure 2). NGS reads with human sequences removed have been deposited in the Sequence Read Archive (BioProject accession no. PRJNA282046; SRP accession no. SRP057614). The 3 CHIKV genome sequences have been deposited in GenBank (accession nos. KR264949–KR264951).
References
- Leparc-Goffart I, Nougairede A, Cassadou S, Prat C, de Lamballerie X. Chikungunya in the Americas. Lancet. 2014;383:514. DOIPubMedGoogle Scholar
- Halstead SB. Reappearance of chikungunya, formerly called dengue, in the Americas. Emerg Infect Dis. 2015;21:557–61.PubMedGoogle Scholar
- Appassakij H, Khuntikij P, Kemapunmanus M, Wutthanarungsan R, Silpapojakul K. Viremic profiles in asymptomatic and symptomatic chikungunya fever: a blood transfusion threat? Transfusion. 2013;53:2567–74. DOIPubMedGoogle Scholar
- Hoad VC, Speers DJ, Keller AJ, Dowse GK, Seed CR, Lindsay MD, First reported case of transfusion-transmitted Ross River virus infection. Med J Aust. 2015;202:267–9. DOIPubMedGoogle Scholar
- Greninger AL, Chen EC, Sittler T, Scheinerman A, Roubinian N, Yu G, A metagenomic analysis of pandemic influenza A (2009 H1N1) infection in patients from North America. PLoS ONE. 2010;5:e13381.PubMedGoogle Scholar
- Chiu CY, Rouskin S, Koshy A, Urisman A, Fischer K, Yagi S, Microarray detection of human parainfluenzavirus 4 infection associated with respiratory failure in an immunocompetent adult. Clin Infect Dis. 2006;43:e71–6. DOIPubMedGoogle Scholar
- Pfeffer M, Linssen B, Parke MD, Kinney RM. Specific detection of chikungunya virus using a RT-PCR/nested PCR combination. J Vet Med B Infect Dis Vet Public Health. 2002;49:49–54. DOIPubMedGoogle Scholar
- Stramer SL, Linnen JM, Carrick JM, Foster GA, Krysztof DE, Zou S, Dengue viremia in blood donors identified by RNA and detection of dengue transfusion transmission during the 2007 dengue outbreak in Puerto Rico. Transfusion. 2012;52:1657–66. DOIPubMedGoogle Scholar
- Stramer SL, Dodd RY, Chiu CY. Advances in testing technology to ensure transfusion safety–NAT and beyond. [ISBT Science Series]. Vox Sang. 2015;10(Suppl 1):55–64. DOIGoogle Scholar
- Naccache SN, Federman S, Veeraraghavan N, Zaharia M, Lee D, Samayoa E, A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res. 2014;24:1180–92. DOIPubMedGoogle Scholar
- Lanciotti RS, Valadere AM. Transcontinental movement of Asian genotype chikungunya virus. Emerg Infect Dis. 2014;20:1400–2. DOIPubMedGoogle Scholar
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. DOIPubMedGoogle Scholar
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. DOIPubMedGoogle Scholar
- Gallian P, de Lamballerie X, Salez N, Piorkowski G, Richard P, Paturel L, Prospective detection of chikungunya virus in blood donors, Caribbean 2014. Blood. 2014;123:3679–81. DOIPubMedGoogle Scholar
- Farrugia A, Kreil TR. Reflections on the emergence of chikungunya virus in the United States: time to revisit a successful paradigm for the safety of blood-derived therapies. Transfusion. 2015;55:224–6. DOIPubMedGoogle Scholar