Volume 22, Number 10—October 2016
Research
Whole-Genome Characterization of Epidemic Neisseria meningitidis Serogroup C and Resurgence of Serogroup W, Niger, 2015
Figure 3
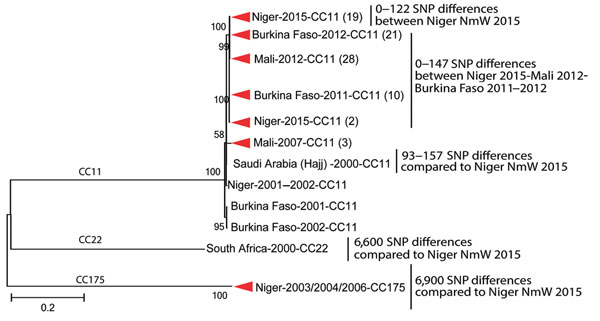
Figure 3. Phylogenetic tree of a subset of the Neisseria meningitidis serogroup W (NmW) isolates, labeled with country of origin, year of isolation, and clonal complex (CC). Clades comprising isolates from a single country and year are collapsed, with the isolate count in parentheses. Internal nodes are labeled with bootstrap values, and the number of single nucleotide polymorphisms (SNPs) distinguishing different groups is provided at right. The scale bar is based on the 11,324 positions in the core SNP matrix and indicates nucleotide substitutions per site.
1These first authors contributed equally to this article.
2Members of the Niger Response Team who contributed to this study are listed at the end of this article.
Page created: September 20, 2016
Page updated: September 20, 2016
Page reviewed: September 20, 2016
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.