Volume 22, Number 11—November 2016
Research
Global Escherichia coli Sequence Type 131 Clade with blaCTX-M-27 Gene
Figure 3
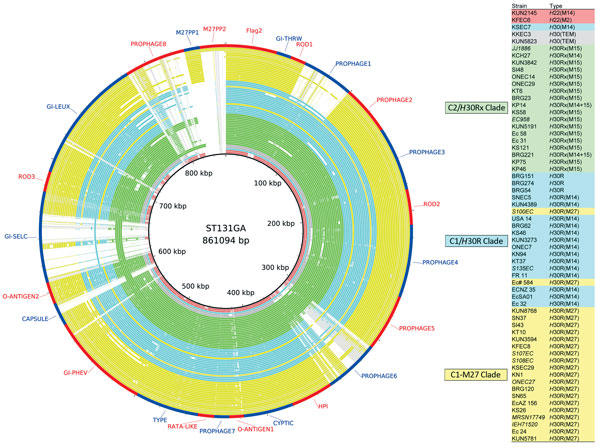
Figure 3. Genome similarities to the Escherichia coli sequence type (ST) 131 genomic islands and the C1-M27 clade–specific region. Rings drawn by BRIG show the presence of these regions. Colored segments indicate >90% similarity and gray segments indicate >70% similarity by BLAST comparison between the regions of interest and each genome. Extended-spectrum β-lactamase types are indicated in parentheses of Type column. The regions from Flag2 to GI-lueX were found in EC958, the prophage 8 region was found in JJ1886, and the M27PP1 and M27PP2 were found as the C1-M27 clade–specific regions in this study. Prophage 6, capsule, GI-selC, and prophage 8 regions were present in some C2/H30Rx isolates but were absent in C1/H30R isolates.
Page created: October 18, 2016
Page updated: October 18, 2016
Page reviewed: October 18, 2016
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.