Volume 22, Number 2—February 2016
Dispatch
The Merits of Malaria Diagnostics during an Ebola Virus Disease Outbreak
Cite This Article
Citation for Media
Abstract
Malaria is a major public health concern in the countries affected by the Ebola virus disease epidemic in West Africa. We determined the feasibility of using molecular malaria diagnostics during an Ebola virus disease outbreak and report the incidence of Plasmodium spp. parasitemia in persons with suspected Ebola virus infection.
The Ebola virus disease (EVD) epidemic occurring in West Africa is unprecedented in its duration and scale; as of October 28, 2015, a total of 28,575 suspected, probable, and confirmed cases, including 11,313 deaths, have been reported (1). Healthcare workers have been severely affected, and the epidemic has resulted in an almost complete breakdown of the public health infrastructure that undoubtedly resulted in many deaths from otherwise treatable conditions and diseases. Particularly concerning are the effects of lapses in childhood vaccination, antenatal and emergency obstetric care, HIV treatment, and malaria control (2–7).
Malaria is a major public health concern in Guinea, Liberia, and Sierra Leone, the 3 countries most affected by the EVD epidemic. In 2012, these countries accounted for ≈9 million malaria cases and 30,566 associated deaths (8). Symptoms of Ebola virus (EBOV) infection and malaria overlap to a great extent, especially early during the course of disease; fever, headache, chills, and vomiting are observed frequently in both diseases. Malaria transmission occurs year-round in Liberia. Therefore, it is recommended that every patient with suspected malaria receive treatment for presumptive malaria when they first seek medical care at an Ebola treatment unit (ETU) or triage point (9,10). A diagnostic test to detect Plasmodium spp. parasitemia was implemented in the joint Centers for Disease Control and Prevention–National Institutes of Health (CDC–NIH) diagnostic laboratory located at the Eternal Love Winning Africa (ELWA) campus in Monrovia, Liberia.
Plasmodium spp. parasitemia can be detected by using rapid diagnostic tests (RDTs), light microscopy, or PCR. Of these methods, PCR is the most sensitive (0.004 parasites/μL) (11), and light microscopy is the reference diagnosis standard (5–10 parasites/μL). Both methods require highly qualified personnel and, in the case of PCR, specialized equipment to perform the test and analyze the results. An RDT produces results quickly and is simple to use, but it is the least sensitive method (>100 parasites/μL) (12), and most RDTs detect only P. falciparum. Light microscopy analysis poses difficulties in an EVD outbreak because of the required handling of infectious material and the need for proper personal protective equipment, so this diagnostic service usually is discontinued during EVD outbreaks. Therefore, because all blood samples submitted to the ELWA laboratory were analyzed by real-time quantitative reverse transcription PCR (qRT-PCR) for the presence of EBOV, we chose to screen for Plasmodium spp. parasitemia also by using qRT-PCR. Here we determine the feasibility of using molecular Plasmodium spp. diagnostics during an EVD outbreak and report the incidence of Plasmodium spp. parasitemia in persons with suspected EBOV infection.
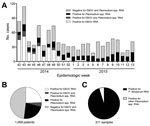
During October 12, 2014–March 28, 2015, samples collected from 1,058 persons in Liberia with suspected EBOV infection were submitted to the CDC–NIH ELWA laboratory. The samples used for this research were collected for public health surveillance and not human subjects research, so institutional review board review and approval were not required. Early during the study period (October–November), most of the patients from whom samples were collected received a diagnosis of EBOV infection (Figure 1, panel A); toward the end, however, few cases of EBOV infection were diagnosed in Liberia. The number of patients who received a diagnosis of Plasmodium spp. parasitemia remained stable over time; thus, despite the lack of positive EBOV test results in the final months of the study period, overall, 40%–60% of patients each week received a diagnosis of EBOV infection, Plasmodium spp. parasitemia, or both (Figure 1, panel A). Of 1,058 samples tested, 259 (24.5%) were positive for EBOV alone, 243 (23%) were positive for Plasmodium spp. alone, and 47 (4.4%) were positive for both (Figure 1, panel B). Of 311 Plasmodium-positive samples that were further analyzed, 296 (95%) were positive for P. falciparum (Figure 1, panel C), confirming that P. falciparum was the main Plasmodium species causing parasitemia in our cohort.
The cycle threshold (Ct) values observed in the Plasmodium spp. qRT-PCR results are a proxy for parasite load (Figure 2). A high Ct value corresponds to a low-level parasitemia; the lower the Ct value, the higher the number of Plasmodium parasites detected. All patients in the cohort were triaged as having suspected EBOV infection, and thus all had clinical symptoms that might have been caused by EBOV, Plasmodium spp., or a different pathogen. Of note, significantly fewer Plasmodium parasites were detected in patients with EBOV infection than in patients who were not infected with EBOV (average Plasmodium Ct 24.7 vs. 20.37; p<0.01 by unpaired Student t-test), likely because clinical symptoms in these patients were caused by the EBOV infection rather than malaria. Some of the patients in our cohort with Plasmodium spp. parasitemia might have been asymptomatic carriers, especially those with a low-level parasitemia.
One could argue that because all patients with febrile illness who were seen at many of the ETUs were given antimalarial treatment when they first sought medical care, providing laboratory testing for Plasmodium spp. parasitemia is not useful. Presumptive artemisinin-based combination treatment is recommended for all patients seen at ETUs, followed by prompt malaria diagnostic testing so that appropriate measures can be taken if oral treatment cannot be sustained because of clinical symptoms (9,10). Moreover, mathematical models predict a large increase in malaria in the epidemic region because of lapses in malaria control (7). Therefore, Plasmodium spp. parasitemia testing of all febrile patients seen at healthcare facilities could be used to determine whether this increase is indeed occurring so that countermeasures can be scaled up accordingly.
Differential diagnostic testing should be expanded to identify the cause of disease in the patients in our cohort whose samples tested negative for EBOV and Plasmodium spp., as was attempted in 1 laboratory in Sierra Leone (15). However, this kind of testing was difficult for several reasons. First, during the height of the EVD epidemic in Liberia, most laboratories were already working close to capacity while testing for EBOV alone. Testing for additional pathogens would have required the allocation of additional resources, including equipment and personnel. Second, it might not be possible to perform diagnostics using a whole blood sample for all the pathogens of differential diagnostic importance that can cause signs and symptoms similar to those of EVD (e.g., typhoid, bacterial sepsis, shigellosis, cholera, leptospirosis, dengue fever, rickettsioses, relapsing fever, meningitis, viral hepatitis, influenza, Lassa fever). Third, each of these pathogens likely is the cause of disease in only a small subset of febrile patients. Last, the prevalence of these pathogens likely differs in each outbreak region. Because broad-spectrum antimicrobial drugs and antimalarial drugs are routinely administered to all patients seen at an ETU, consideration for additional testing should focus on endemic pathogens that would not respond to these treatments.
By using PCR-based detection for Plasmodium spp. parasitemia, the need for additional handling of clinical specimens possibly infected with EBOV was eliminated. Thus, the addition of PCR-based diagnostic tests to detect Plasmodium spp. does not pose an additional safety risk to laboratory staff. Also, it is less time-consuming to add additional PCR reactions to a PCR run than to separately perform microscopy or an RDT on each sample, and these additions would add only ≈15 minutes to the overall time needed from sample submission to reporting of results. Moreover, if a multiplex approach is used, the additions would not require extra time or reagents. Taken together, our findings suggest that PCR-based testing for Plasmodium spp. parasitemia can be implemented easily and safely in laboratories performing EBOV diagnostics to assist with case-patient management during EVD outbreaks in malaria-endemic areas.
Dr. de Wit is a research fellow in the Disease Modeling and Transmission Section of the Laboratory of Virology, Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, NIH, Hamilton, Montana. During the Ebola outbreak in West Africa, she spent 4 months working in the CDC–NIH ELWA field laboratory in Monrovia, Liberia, providing Ebola diagnostics to several ETUs and other healthcare facilities.
Acknowledgments
The authors would like to thank Mulbah Jallah, Monrovia, Liberia, for his excellent support and assistance in operating the laboratory at ELWA, as well as Kay Menk and Dawn Clifton for logistical support. We further acknowledge the World Health Organization headquarters in Geneva, Switzerland; the World Health Organization Regional Office for Africa; the Liberia Ministry of Health and Social Welfare; CDC; NIH; and ELWA. Last, we would like to thank the people of Liberia for their hospitality and cooperation.
This work was funded in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, NIH.
References
- World Health Organization. Ebola situation report. 2015 October 28 [cited 2015 Oct 28]. http://apps.who.int/iris/bitstream/10665/191299/1/ebolasitrep_28Oct2015_eng.pdf?ua=1
- Plucinski MM, Guilavogui T, Sidikiba S, Diakité N, Diakité S, Dioubaté M, Effect of the Ebola-virus-disease epidemic on malaria case management in Guinea, 2014: a cross-sectional survey of health facilities. Lancet Infect Dis. 2015;15:1017–23 .DOIPubMedGoogle Scholar
- Mobula ML, Brown CA, Burnham G, Phelps BR. Need for reinforced strategies to support delivery of HIV clinical services during the Ebola outbreak in Guinea, Liberia, and Sierra Leone. Disaster Med Public Health Prep. 2015;9:522–6 .DOIPubMedGoogle Scholar
- Takahashi S, Metcalf CJ, Ferrari MJ, Moss WJ, Truelove SA, Tatem AJ, Reduced vaccination and the risk of measles and other childhood infections post-Ebola. Science. 2015;347:1240–2 .DOIPubMedGoogle Scholar
- United Nations Population Fund. Ebola wiping out gains in safe motherhood [press release]. 16 October 2014 [cited 2015 Oct 28]. http://www.unfpa.org/news/ebola-wiping-out-gains-safe-motherhood
- Walker PG, White MT, Griffin JT, Reynolds A, Ferguson NM, Ghani AC. Malaria morbidity and mortality in Ebola-affected countries caused by decreased health-care capacity, and the potential effect of mitigation strategies: a modelling analysis. Lancet Infect Dis. 2015;15:825–32 .DOIPubMedGoogle Scholar
- World Health Organization. World malaria report 2014 [cited 2015 Oct 28]. http://www.who.int/malaria/publications/world_malaria_report_2014/report/en
- Centers for Disease Control and Prevention. Recommendations for managing and preventing cases of malaria in areas with Ebola. 2015 [cited 2015 Oct 28]. http://www.cdc.gov/vhf/ebola/outbreaks/malaria-cases.html
- Sterk E. Filovirus haemorrhagic fever guideline. Barcelona, Spain: Médecins Sans Frontières; 2008.
- Benito A, Rubio JM. Usefulness of seminested polymerase chain reaction for screening blood donors at risk for malaria in Spain. Emerg Infect Dis. 2001;7:1068 .DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. DPDx: laboratory identification of parasitic diseases of public health concern. 2015 [cited 2015 Oct 28]. http://www.cdc.gov/dpdx/diagnosticProcedures/blood/antigendetection.html
- Lee MA, Tan CH, Aw LT, Tang CS, Singh M, Lee SH, Real-time fluorescence-based PCR for detection of malaria parasites. J Clin Microbiol. 2002;40:4343–5 .DOIPubMedGoogle Scholar
- Shokoples SE, Ndao M, Kowalewska-Grochowska K, Yanow SK. Multiplexed real-time PCR assay for discrimination of Plasmodium species with improved sensitivity for mixed infections. J Clin Microbiol. 2009;47:975–80 .DOIPubMedGoogle Scholar
- O’Shea MK, Clay KA, Craig DG, Matthews SW, Kao RL, Fletcher TE, Diagnosis of febrile illnesses other than Ebola virus disease at an Ebola treatment unit in Sierra Leone. Clin Infect Dis. 2015;61:795–8 .DOIPubMedGoogle Scholar
Figures
Cite This Article1Current affiliation: University of Saskatchewan, Saskatoon, Saskatchewan, Canada.
2Current affiliation: Public Health Agency of Canada, Winnipeg, Manitoba, Canada.
3Current affiliation: Friedrich-Loeffler-Institut, Greifswald-Insel Riems, Germany.
Table of Contents – Volume 22, Number 2—February 2016
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Vincent J. Munster, Laboratory of Virology, National Institutes of Health, 903 S 4th St, Hamilton, MT 59840, USA
Top