Volume 23, Number 10—October 2017
Dispatch
Molecular Tracing to Find Source of Protracted Invasive Listeriosis Outbreak, Southern Germany, 2012–2016
Figure 1
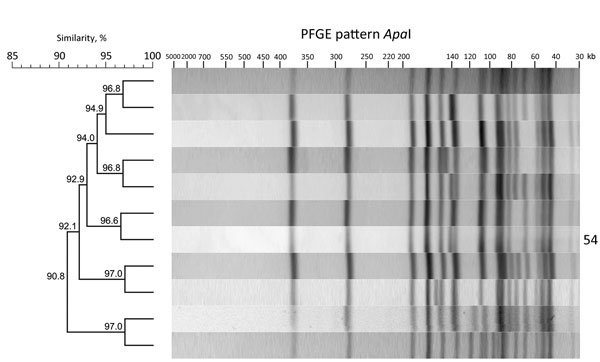
Figure 1. ApaI restriction enzyme analysis of Listeria monocytogenes outbreak strain and isolates with >90% similarity to the outbreak strain, southern Germany, 2012–2016. We performed molecular subtyping in line with the PulseNet standardized PFGE protocol for L. monocytogenes (8) and the standard operating procedures of the European Union Reference Laboratory for L. monocytogenes (9) to ensure interlaboratory comparability of the results. We analyzed PFGE patterns using BioNumerics software version 7.5 (Applied Maths, Sint-Martens-Latem, Belgium). Dendrogram indicates percentage similarity between the ApaI PFGE pattern of the outbreak strain and that of the other closely related isolates. Outbreak strain PFGE pattern is labeled with the number 54. PFGE, pulsed-field gel electrophoresis.
References
- Farber JM, Peterkin PI. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511.PubMedGoogle Scholar
- Schuchat A, Swaminathan B, Broome CV. Epidemiology of human listeriosis. Clin Microbiol Rev. 1991;4:169–83. DOIPubMedGoogle Scholar
- Robert Koch-Institute. Infektionsepidemiologisches Jahrbuch meldepflichtiger Krankheiten für 2016. Berlin: The Institute; 2017 [cited 2017 Mar 1]. http://www.rki.de/jahrbuch
- Preußel K, Milde-Busch A, Schmich P, Wetzstein M, Stark K, Werber D. Risk factors for sporadic non-pregnancy associated listeriosis in Germany—immunocompromised patients and frequently consumed ready-to-eat products. PLoS One. 2015;10:e0142986. DOIPubMedGoogle Scholar
- Werber D, Hille K, Frank C, Dehnert M, Altmann D, Müller-Nordhorn J, et al. Years of potential life lost for six major enteric pathogens, Germany, 2004-2008. Epidemiol Infect. 2013;141:961–8. DOIPubMedGoogle Scholar
- Ruppitsch W, Prager R, Halbedel S, Hyden P, Pietzka A, Huhulescu S, et al. Ongoing outbreak of invasive listeriosis, Germany, 2012 to 2015. Euro Surveill. 2015;20:pii=30094. DOIPubMedGoogle Scholar
- Ruppitsch W, Pietzka A, Prior K, Bletz S, Fernandez HL, Allerberger F, et al. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J Clin Microbiol. 2015;53:2869–76. DOIPubMedGoogle Scholar
- Halpin JL, Garrett NM, Ribot EM, Graves LM, Cooper KL. Re-evaluation, optimization, and multilaboratory validation of the PulseNet-standardized pulsed-field gel electrophoresis protocol for Listeria monocytogenes. Foodborne Pathog Dis. 2010;7:293–8. DOIPubMedGoogle Scholar
- Félix B, Dao TT, Lombard B, Asséré A, Brisabois A, Roussel S. The use of pulsed field gel electrophoresis in Listeria monocytogenes sub-typing—harmonization at the European Union level. In: Magdeldin S, editor. Gel electrophoresis—principles and basics. Rijeka (Croatia): In Tech; 2012. p. 241–54.