Volume 23, Number 10—October 2017
Dispatch
Berlin Squirrelpox Virus, a New Poxvirus in Red Squirrels, Berlin, Germany
Figure 2
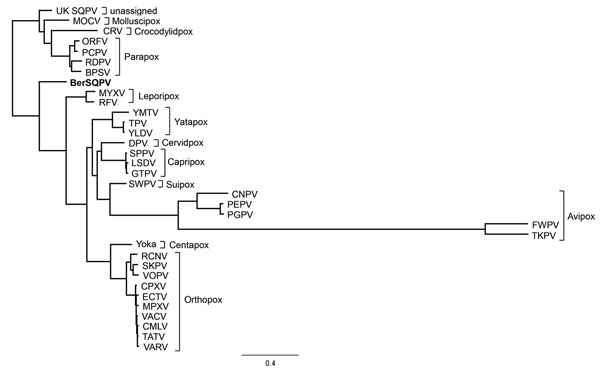
Figure 2. Phylogenetic position of BerSQPV (bold) from a red squirrel in Berlin, Germany, within the Chordopoxvirinae. We used MAFFT (12) to perform multiple alignments of all complete genome sequences within a species of the Chordopoxvirinae subfamily available in GenBank. The minimum pairwise identity found within any of these intraspecies alignments was 79.1%; the maximum pairwise identity of BerSQPV with any chordopoxvirus genome available was 47%. Because of this extreme difference in minimum pairwise identities, we selected individual prototype genomes for each species and the viruses with highest identity to BerSQPV for phylogenetic analysis (as indicated in figure). We performed a multiple alignment of these representative sequences with the BerSQPV genome and removed low-quality regions from the alignment using Gblocks version 0.91(13), yielding a stripped alignment of 52,563 gap-free positions. The maximum-likelihood tree was then calculated using PhyML(14) (general time reversible plus gamma, 4 substitution rate categories, no invariable sites, BEST topology search, χ2-based parametric branch supports). Scale bar indicates nucleotide substitutions per site. BPSV, bovine papular stomatitis virus BV-AR02 (NC_005337); CMLV, camelpox virus CMS (AY009089); CNPV, canarypox virus Wheatley C93 (NC_005309); CPXV, cowpox virus Brighton Red (AF482758); CRV, Nile crocodilepox virus (NC_008030); DPV, deerpox virus W-848–83 (NC_006966); ECTV, ectromelia virus Moscow (AF012825); FWPV, fowlpox virus NVSL (NC_002188); GTPV, goatpox virus Pellor (NC_004003); LSDV, lumpy skin disease virus NI-2490 (NC_003027); MOCV, Molluscum contagiosum virus subtype 1 (NC_001731); MPXV, monkeypox virus Zaire-96-I-16 (AF380138); MYXV, myxoma virus Lausanne (NC_001132); ORFV, Orf virus OV-SA00 (NC_005336); PCPV, pseudocowpox virus VR634 (NC_013804); PEPV, penguinpox virus (KJ859677); PGPV, pigeonpox virus FeP2 (NC_024447); RCNV, raccoonpox virus Herman (NC_027213); RDPV, red deer pox virus (KM502564); RFV, rabbit fibroma virus Kasza (AF170722); SKPV, skunkpox virus (KU749310); SPPV, sheeppox virus 17077–99 (NC_004002); UK SQPV, squirrel poxvirus Red squirrel UK (HE601899); SWPV, swinepox virus 17077–99 (NC_003389); TATV, taterapox virus Dahomey 1968 (NC_008291); TKPV, turkeypox virus HU1124/2011 (KP728110); TPV, tanapox virus (EF420156); FukVACV, vaccinia virus Copenhagen (M35027); VARV, variola major virus Bangladesh-1975 (L22579); VPXV, volepox virus (KU749311); YLDV, Yaba-like disease virus (NC_002642); YMTV, Yaba monkey tumor virus (NC_005179); Yoka, Yokapox virus (NC_015960)].
References
- Sainsbury AW, Deaville R, Lawson B, Cooley WA, Farelly SS, Stack MJ, et al. Poxviral disease in red squirrels Sciurus vulgaris in the UK: spatial and temporal trends of an emerging threat. EcoHealth. 2008;5:305–16. DOIPubMedGoogle Scholar
- Scott AC, Keymer IF, Labram J. Parapoxvirus infection of the red squirrel (Sciurus vulgaris). Vet Rec. 1981;109:202. DOIPubMedGoogle Scholar
- McInnes CJ, Wood AR, Thomas K, Sainsbury AW, Gurnell J, Dein FJ, et al. Genomic characterization of a novel poxvirus contributing to the decline of the red squirrel (Sciurus vulgaris) in the UK. J Gen Virol. 2006;87:2115–25. DOIPubMedGoogle Scholar
- Himsworth CG, Musil KM, Bryan L, Hill JE. Poxvirus infection in an American red squirrel (Tamiasciurus hudsonicus) from northwestern Canada. J Wildl Dis. 2009;45:1143–9. DOIPubMedGoogle Scholar
- Obon E, Juan-Sallés C, McInnes CJ, Everest DJ. Poxvirus identified in a red squirrel (Sciurus vulgaris) from Spain. Vet Rec. 2011;168:86. DOIPubMedGoogle Scholar
- Kurth A, Nitsche A. Detection of human-pathogenic poxviruses. Methods Mol Biol. 2010;665:257–78. DOIPubMedGoogle Scholar
- Li Y, Meyer H, Zhao H, Damon IK. GC content-based pan-pox universal PCR assays for poxvirus detection. J Clin Microbiol. 2010;48:268–76. DOIPubMedGoogle Scholar
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. DOIPubMedGoogle Scholar
- Tausch SH, Renard BY, Nitsche A, Dabrowski PW. RAMBO-K: rapid and sensitive removal of background sequences from next generation sequencing data. PLoS One. 2015;10:e0137896. DOIPubMedGoogle Scholar
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. DOIPubMedGoogle Scholar
- Darby AC, McInnes CJ, Kjær KH, Wood AR, Hughes M, Martensen PM, et al. Novel host-related virulence factors are encoded by squirrelpox virus, the main causative agent of epidemic disease in red squirrels in the UK. PLoS One. 2014;9:e96439. DOIPubMedGoogle Scholar
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. DOIPubMedGoogle Scholar
- Talavera G, Castresana J, Kjer K, Page R, Sullivan J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–77. DOIPubMedGoogle Scholar
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. DOIPubMedGoogle Scholar
- Himsworth CG, McInnes CJ, Coulter L, Everest DJ, Hill JE. Characterization of a novel poxvirus in a North American red squirrel (Tamiasciurus hudsonicus). J Wildl Dis. 2013;49:173–9. DOIPubMedGoogle Scholar
1These authors contributed equally to this article.