Volume 23, Number 12—December 2017
Dispatch
Phylogenetic Characterization of Crimean-Congo Hemorrhagic Fever Virus, Spain
Figure
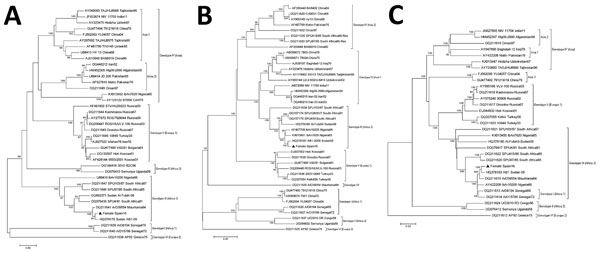
Figure. Phylogenetic analysis of Crimean-Congo hemorrhagic fever virus from patient in Spain, 2016, compared with reference sequences. A) Small segment (1,450 bp); B) medium segment (4,497 bp); C) large segment (11,829 bp). Trees were generated with the neighbor-joining method with Kimura 2-parameter distances by using MEGA version 5.1 (http://www.megasoftware.net). Bootstrap confidence limits were calculated on the basis of 1,000 replicates; numbers on branches indicate bootstrap results. Triangles indicate newly sequenced strain from Spain described in this article; other sequences are named by GenBank accession number, strain, geographic origin, and sampling year. Genotypes at right are named according to Carrol et al. (4); brackets indicate equivalent group nomenclature according to Chamberlain et al. (1). Roman numerals indicate geographic locations: I, West Africa (Africa 1); II, Central Africa (Africa 2); III, South and West Africa (Africa 3); IV, Middle East/Asia, divided into 2 groups, Asia 1 and Asia 2 (1); V, Europe/Turkey (Europe 1); VI, Greece (Europe 2). Scale bars indicate nucleotide substitutions per site.
References
- Chamberlain J, Cook N, Lloyd G, Mioulet V, Tolley H, Hewson R. Co-evolutionary patterns of variation in small and large RNA segments of Crimean-Congo hemorrhagic fever virus. J Gen Virol. 2005;86:3337–41. DOIPubMedGoogle Scholar
- Deyde VM, Khristova ML, Rollin PE, Ksiazek TG, Nichol ST. Crimean-Congo hemorrhagic fever virus genomics and global diversity. J Virol. 2006;80:8834–42. DOIPubMedGoogle Scholar
- Anagnostou V, Papa A. Evolution of Crimean-Congo hemorrhagic fever virus. Infect Genet Evol. 2009;9:948–54. DOIPubMedGoogle Scholar
- Carroll SA, Bird BH, Rollin PE, Nichol ST. Ancient common ancestry of Crimean-Congo hemorrhagic fever virus. Mol Phylogenet Evol. 2010;55:1103–10. DOIPubMedGoogle Scholar
- Aradaib IE, Erickson BR, Karsany MS, Khristova ML, Elageb RM, Mohamed ME, et al. Multiple Crimean-Congo hemorrhagic fever virus strains are associated with disease outbreaks in Sudan, 2008-2009. PLoS Negl Trop Dis. 2011;5:e1159. DOIPubMedGoogle Scholar
- Singh P, Chhabra M, Sharma P, Jaiswal R, Singh G, Mittal V, et al. Molecular epidemiology of Crimean-Congo haemorrhagic fever virus in India. Epidemiol Infect. 2016;15:1–4.PubMedGoogle Scholar
- Hoogstraal H. The epidemiology of tick-borne Crimean-Congo hemorrhagic fever in Asia, Europe, and Africa. J Med Entomol. 1979;15:307–417. DOIPubMedGoogle Scholar
- Bente DA, Forrester NL, Watts DM, McAuley AJ, Whitehouse CA, Bray M. Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res. 2013;100:159–89. DOIPubMedGoogle Scholar
- Filipe AR, Calisher CH, Lazuick J. Antibodies to Congo-Crimean haemorrhagic fever, Dhori, Thogoto and Bhanja viruses in southern Portugal. Acta Virol. 1985;29:324–8.PubMedGoogle Scholar
- Estrada-Peña A, Palomar AM, Santibáñez P, Sánchez N, Habela MA, Portillo A, et al. Crimean-Congo hemorrhagic fever virus in ticks, Southwestern Europe, 2010. Emerg Infect Dis. 2012;18:179–80. DOIPubMedGoogle Scholar
- Palomar AM, Portillo A, Santibáñez P, Mazuelas D, Arizaga J, Crespo A, et al. Crimean-Congo hemorrhagic fever virus in ticks from migratory birds, Morocco. Emerg Infect Dis. 2013;19:260–3. DOIPubMedGoogle Scholar
- Negredo A, de la Calle-Prieto F, Palencia-Herrejón E, Mora-Rillo M, Astray-Mochales J, Sánchez-Seco MP, et al.; Crimean Congo Hemorrhagic Fever@Madrid Working Group. Autochthonous Crimean-Congo Hemorrhagic Fever in Spain. N Engl J Med. 2017;377:154–61. DOIPubMedGoogle Scholar
- Blackley DJ, Wiley MR, Ladner JT, Fallah M, Lo T, Gilbert ML, Gregory C, et al. Reduced evolutionary rate in reemerged Ebola virus transmission chains. Sci Adv. 2016;29;2:e1600378.
1These authors contributed equally to this article.