Volume 23, Number 2—February 2017
Dispatch
Nosocomial Infections with IMP-19−Producing Pseudomonas aeruginosa Linked to Contaminated Sinks, France
Cite This Article
Citation for Media
Abstract
We isolated IMP-19–producing Pseudomonas aeruginosa from 7 patients with nosocomial infections linked to contaminated sinks in France. We showed that blaIMP-19 was located on various class 1 integrons among 8 species of gram-negative bacilli detected in sinks: P. aeruginosa, Achromobacter xylosoxidans, A. aegrifaciens, P. putida, Stenotrophomonas maltophilia, P. mendocina, Comamonas testosteroni, and Sphingomonas sp.
Acquired metallo-β-lactamases (MBLs) belong to the families IMP, VIM, NDM, SPM, GIM, SIM, DIM, KHM, TMB, FIM, and AIM (1). IMP and VIM are the most common families. MBLs have been reported worldwide among Pseudomonas aeruginosa isolates (2). Therapeutic options for infected patients are severely limited because isolates are resistant to many classes of antimicrobial drugs. Genes for MBLs are found mostly in class 1 integrons, which carry additional drug resistance genes. To date, 33 of the 51 known IMP variants have been detected in P. aeruginosa; there has been only 1 report of an IMP-19 producer (3). This MBL is widespread among Acinetobacter spp. in Japan and has also been reported in Achromobacter xylosoxidans (4,5).
During 2009–2016, infections with IMP-19–producing P. aeruginosa isolates were detected in 7 patients in the Hematology Department of University Hospital François Mitterrand, a 1,600-bed hospital in Dijon, France. We describe these infections and report results of environmental investigations.
The hematology department of the hospital contains a 15-bed conventional unit and a 9-bed protective isolation unit. At the entrances of rooms in the conventional unit, there is a hand hygiene sink (for staff and visitors) and a bathroom in a separate area (shower stall, sink, and toilets). In the protective isolation unit, air is filtered through a laminar flow system, and a sink and toilets are located next to each bed (distance 1.5 m). The ceramic sinks have no counter space, and drains are made of stainless steel. All faucets in the department are hand-operated and provided with antibacterial filters (0.22 μm). Surfaces are cleaned daily (once in the conventional unit) and twice in the protective isolation unit with a solution containing quaternary ammonium compounds (0.25% didecyldimethylammonium chloride).
During 2009–2016, a total of 7 patients (P1–P7) in the department were infected or colonized by IMP-19–producing P. aeruginosa, which were isolated from blood samples (2 patients), urine samples (2 patients), throat swab samples (2 patients), and a central venous catheter (1 patient). All patients underwent throat, urine, and feces sampling at admission and were free of P. aeruginosa, thus indicating nosocomial acquisition. All isolates were resistant to ceftazidime, imipenem, meropenem, doripenem, ciprofloxacin, and most aminoglycosides; 4 isolates were susceptible to piperacillin and 3 to amikacin.
We conducted an environmental investigation in the hospital (Figure 1). More than 100 environmental samples were obtained when no patients were colonized (except for samples collected in room 32 a few hours after patient P7 had been transferred to an intensive care unit). Water samples were collected from different faucets (nursing room, medication preparation rooms, and rooms of some patients). First-catch lukewarm water (500 mL) was collected in sterile bottles containing 20 mg/L sodium thiosulfate and concentrated by filtration (0.45-μm membrane filters). All sinks and shower drains were sampled by rotating a cotton swab inserted through the drain. Toilets were sampled with swabs inserted under the rim of the toilet bowl.
We plated samples from filters (for water) and swab specimens (from sinks) on Drigalski medium containing ceftazidime (32 mg/L). When a positive culture was observed after 48 hours of incubation, a PCR specific for blaIMP-19 was performed for the culture mixture. For samples with a positive PCR result, we then plated a dilution of the culture mixture on Drigalski medium containing ceftazidime (32 mg/L) and imipenem (16 mg/L) to obtain isolated colonies, which we further subjected to identification and confirmation of IMP-19 production.
Many resistant organisms were detected in these environmental samples. We used mass spectrometry and nrdA gene sequencing to identify for Achromobacter isolates (6). blaIMP-19 and integrons were detected as described (7,8). We used pulsed-field gel electrophoresis after XbaI digestion to genotype P. aeruginosa isolates (9). Pulsotypes were compared by calculating the Dice correlation coefficient with DendroUPGMA software (http://genomes.urv.cat/UPGMA/).
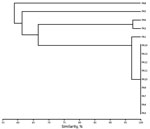
The 7 clinical isolates belonged to 3 distinct genotypes A, B, and C (Table; Figure 2). We detected environmental IMP-19–producing isolates belonging to 8 species of gram-negative nonfermenting bacilli: P. aeruginosa, Achromobacter xylosoxidans, A. aegrifaciens, P. putida, Stenotrophomonas maltophilia, P. mendocina, Comamonas testosteroni, and Sphingomonas sp. Of the 7 environmental isolates of P. aeruginosa we identified, 6 belonged to the same genotype as clinical isolates (genotype A).
The blaIMP-19 gene was located in various integrons, mainly on a sul-type class 1 integron and A Tn402-like class 1 integron. In these integrons, blaIMP-19 was associated with different gene cassettes, including aac(6′)-Ib, aadA13, aadB, or fused qacG-aac(6′)-Ib (Figure 3). Few isolates had several copies of blaIMP-19 located on integrons of different structures (Table).
The incidence of MBL producers among imipenem-resistant P. aeruginosa in France is low compared with incidences for other countries (2,10). Reports of outbreaks are scarce and usually involve VIM producers (11,12). Therefore, detection of IMP-19 producers in our hospital was unusual. The long intervals without cases, the absence of any overlap between cases, and genotypic diversity of clinical isolates did not suggest a single common source of infection. These findings prompted us to conduct environmental investigations.
IMP-19 producers were detected in 9 of 15 rooms in the conventional unit and 2 of 9 rooms in the protective isolation unit. These producers were P. aeruginosa and a wide variety of gram-negative nonfermenting bacilli. Most of these producers have little clinical relevance, but they are silent reservoirs for dissemination of blaIMP-19 to major pathogens. The role of these environmental bacterial species in the spread of MBL suggested in previous studies (13,14) is confirmed by our findings.
The diversity of species found and genetic structures involved with blaIMP-19 indicated that the environmental contamination occurred a long time ago. One isolate of IMP-19–producing Aeromonas caviae was found in a patient in the same building in 2006 (7). This phenomenon is probably endemic to our hospital, in which sink drains are not accessible for removal of biofilms without complete dismantling (inappropriate sink design).
Transfer of pathogens from sinks to patients might occur in several ways. Water from faucets is directed straight into the drain, resulting in splashing that can lead to contamination of an area <1 m from the sink (15). Therefore, patients can be contaminated when they brush their teeth, wash their hands, or take a shower. Healthcare personnel are also at risk for hand contamination, which might lead to transfer of pathogens to patients during care.
All patients had >1 stay in rooms that were positive for IMP-19–producing organisms. After patient P7 died of sepsis, all drains in the ward were changed. However, this measure did not eradicate biofilms found in the plumbing system. Because the building tested was 16 years old, it has been decided to completely rebuild the ward in early 2017, paying special attention to water distribution and discharge systems to minimize hospital-acquired infections. In conclusion, our findings might help other hospitals to identify potential reservoirs of carbapenemase-producing bacteria and lead to implementation of rapid control measures to contain outbreaks.
Dr. Amoureux is research scientist in the Bacteriology Department, University Hospital François Mitterrand, Dion, France. Her research interests are epidemiology and resistance of nonfermenting gram-negative bacilli, specifically Achromobacter spp., in immunocompromised patients and those with cystic fibrosis.
Acknowledgments
We thank N. Sixt for performing water analysis and P. Bastable for correcting and editing the manuscript.
This study was supported by the Association Dijonnaise des Bactériologistes.
References
- Zhao WH, Hu ZQ. Acquired metallo-β-lactamases and their genetic association with class 1 integrons and ISCR elements in Gram-negative bacteria. Future Microbiol. 2015;10:873–87. DOIPubMedGoogle Scholar
- Cornaglia G, Giamarellou H, Rossolini GM. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect Dis. 2011;11:381–93. DOIPubMedGoogle Scholar
- Pollini S, Antonelli A, Venturelli C, Maradei S, Veggetti A, Bracco S, et al. Acquisition of plasmid-borne blaIMP-19 gene by a VIM-1-positive Pseudomonas aeruginosa of the sequence type 235 epidemic lineage. J Antimicrob Chemother. 2013;68:722–4. DOIPubMedGoogle Scholar
- Yamamoto M, Nagao M, Matsumura Y, Hotta G, Matsushima A, Ito Y, et al. Regional dissemination of Acinetobacter species harbouring metallo-β-lactamase genes in Japan. Clin Microbiol Infect. 2013;19:729–36. DOIPubMedGoogle Scholar
- Yamamoto M, Nagao M, Hotta G, Matsumura Y, Matsushima A, Ito Y, et al. Molecular characterization of IMP-type metallo-β-lactamases among multidrug-resistant Achromobacter xylosoxidans. J Antimicrob Chemother. 2012;67:2110–3. DOIPubMedGoogle Scholar
- Spilker T, Vandamme P, Lipuma JJ. Identification and distribution of Achromobacter species in cystic fibrosis. J Cyst Fibros. 2013;12:298–301. DOIPubMedGoogle Scholar
- Neuwirth C, Siebor E, Robin F, Bonnet R. First occurrence of an IMP metallo-beta-lactamase in Aeromonas caviae: IMP-19 in an isolate from France. Antimicrob Agents Chemother. 2007;51:4486–8. DOIPubMedGoogle Scholar
- Betteridge T, Partridge SR, Iredell JR, Stokes HW. Genetic context and structural diversity of class 1 integrons from human commensal bacteria in a hospital intensive care unit. Antimicrob Agents Chemother. 2011;55:3939–43. DOIPubMedGoogle Scholar
- Tsakris A, Tassios PT, Polydorou F, Papa A, Malaka E, Antoniadis A, et al. Infrequent detection of acquired metallo-beta-lactamases among carbapenem-resistant Pseudomonas isolates in a Greek hospital. Clin Microbiol Infect. 2003;9:846–51. DOIPubMedGoogle Scholar
- Fournier D, Richardot C, Müller E, Robert-Nicoud M, Llanes C, Plésiat P, et al. Complexity of resistance mechanisms to imipenem in intensive care unit strains of Pseudomonas aeruginosa. J Antimicrob Chemother. 2013;68:1772–80. DOIPubMedGoogle Scholar
- Ambrogi V, Cavalié L, Mantion B, Ghiglia MJ, Cointault O, Dubois D, et al. Transmission of metallo-β-lactamase-producing Pseudomonas aeruginosa in a nephrology-transplant intensive care unit with potential link to the environment. J Hosp Infect. 2016;92:27–9. DOIPubMedGoogle Scholar
- Corvec S, Poirel L, Espaze E, Giraudeau C, Drugeon H, Nordmann P. Long-term evolution of a nosocomial outbreak of Pseudomonas aeruginosa producing VIM-2 metallo-enzyme. J Hosp Infect. 2008;68:73–82. DOIPubMedGoogle Scholar
- Bogaerts P, Bouchahrouf W, Lissoir B, Denis O, Glupczynski Y. IMP-13-producing Pseudomonas monteilii recovered in a hospital environment. J Antimicrob Chemother. 2011;66:2434–5. DOIPubMedGoogle Scholar
- Gilarranz R, Juan C, Castillo-Vera J, Chamizo FJ, Artiles F, Álamo I, et al. First detection in Europe of the metallo-β-lactamase IMP-15 in clinical strains of Pseudomonas putida and Pseudomonas aeruginosa. Clin Microbiol Infect. 2013;19:E424–7. DOIPubMedGoogle Scholar
- Hota S, Hirji Z, Stockton K, Lemieux C, Dedier H, Wolfaardt G, et al. Outbreak of multidrug-resistant Pseudomonas aeruginosa colonization and infection secondary to imperfect intensive care unit room design. Infect Control Hosp Epidemiol. 2009;30:25–33. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleTable of Contents – Volume 23, Number 2—February 2017
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Catherine Neuwirth, Bacteriology Department, University Hospital François Mitterrand, PTB, BP 37013, 21070 Dijon CEDEX, France
Top