Volume 31, Number 8—August 2025
Research Letter
Emergence of Novel Fluoroquinolone Resistance Mutations in Mycoplasma bovis, China, 2008–2023
Abstract
We investigated quinolone resistance in Mycoplasma bovis samples isolated in China during 2008–2023. Sequence type 52 was the dominant genotype; GyrA (S83F/Y) and ParC (S80R) protein double mutations caused high resistance to fluoroquinolones. Increased vigilance and surveillance of M. bovis infections in cattle will be needed to prevent disease.
Diseases in cattle caused by Mycoplasma bovis include bronchopneumonia, mastitis, and arthritis (1,2). M. bovis was first isolated in 1961 (3) and, over the past >6 decades, it has become widespread worldwide. Bovine mycoplasmosis caused by M. bovis is an emerging disease in China. Since the first isolation of M. bovis strains in China’s Hubei region in 2008, those strains have spread rapidly and extensively to most provinces in China (4–6). However, the epidemiologic features of M. bovis in China are unknown. Antimicrobial drugs are currently a critical means of controlling M. bovis infections (7,8). Fluoroquinolones have a substantial bactericidal effect against Mycoplasma spp.; however, their effectiveness has been gradually declining (9,10). Fluoroquinolone resistance in Mycoplasma spp. relies primarily on gene point mutations (7).
To elucidate molecular epidemiologic features of M. bovis in China, we performed a genetic evolutionary analysis of whole-genome sequences from 77 M. bovis isolates collected during 2008–2023 from 16 provinces in China; 34 isolates were identified in this study and 43 isolates were from GenBank (Appendix Table 1). We deposited sequence data for the M. bovis isolates from this study in the National Center for Biotechnology Information BioProject database (https://www.ncbi.nlm.nih.gov/bioproject; accession nos. PRJNA1124599–601).
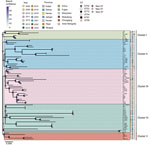
We explored the contribution of genetic factors to fluoroquinolone resistance. We confirmed that sequence type (ST) 52, the primary genotype responsible for the M. bovis infection outbreak in 2008, was the most prevalent genotype in China; however, the topologic structure of the phylogenetic tree classified the 77 isolates into 5 distinct clusters (I–V) (Figure 1). Five of those isolates represented new multilocus sequence typing (MLST) genotypes, which were primarily concentrated in cluster IV (which contained 4 MLST genotypes) (Figure 1; Appendix Figure 1), suggesting that isolates within cluster IV might have undergone rapid genetic changes. During the disease outbreak in 2008, ST53, ST56, and ST72 genotypes were also identified. Although those 3 genotypes were distributed sporadically, they have been isolated only in China and belong to the same clonal complex (CC) 52 as ST52 (Appendix Figure 1), exhibiting a high degree of genetic relatedness. That observation suggests that ST52 underwent genetic variation after spreading extensively in China. ST89 was isolated from cows with pneumonia and mastitis in China during 2018–2019 (Appendix Table 2); however, that genotype does not belong to CC52 (Appendix Figure 1). The isolation of only 3 ST89 strains suggested that strains with other genotypes might be infecting cattle in China.
We analyzed mutations within quinolone resistance–determining regions of the 77 isolated genomes from China. Mutations in those regions occurred primarily in parC and gyrA genes, leading to amino acid changes (Appendix Figure 2). Specifically, the GyrA protein contained S83Y and S83F mutations (Figure 2, panel A), and ParC contained S80R and D84G (Figure 2, panel B). The S80R mutation in ParC is uncommon in M. bovis and has not been reported in China. The binding energies of the GyrA S83F and S83Y and ParC S80R mutants with ciprofloxacin were higher than those for wild-type GyrA and ParC proteins. The mean + SD binding energy increased from −46.115 + 8.72 in wild-type GyrA to −10.242 + 2.892 in the GyrA S83Y mutant (Appendix Table 3). The ParC S80R mutant had considerably higher binding energy than wild-type ParC, increasing from −19.973 + 2.445 in wild-type protein to 26.861 + 5.14 in the mutant. Those mutations led to a decreased and unstable binding capacity of GyrA and ParC with ciprofloxacin.
We investigated the effect of mutations on fluoroquinolone susceptibility of M. bovis. Clinical isolates with the GyrA S83Y/F and ParC S80R double mutations exhibited lower susceptibility to fluoroquinolones than strains that had the GyrA S83F and ParC D84G double mutations (Appendix Table 4, Figure 3), suggesting that S83Y/F in GyrA combined with S80R in ParC conferred high resistance to fluoroquinolones; the S80R ParC mutation appeared to be the main reason for increased fluoroquinolone resistance. Molecular dynamic simulations revealed that residue S80 of M. bovis ParC interacts with enrofloxacin through van der Waals forces (Appendix Figure 4). Strains with GyrA and ParC mutations were mainly concentrated in cluster II (Figure 1), suggesting that cluster II strains are more prone to developing genetic features that confer resistance to fluoroquinolones.
In conclusion, we report that ST52 is the dominant M. bovis genotype circulating in China; however, ST52 strains gradually formed 2 subgroups with dominant genetic variation and fluoroquinolone resistance through widespread dissemination. The double mutation, S83F in GyrA and S80R in ParC, appears to be the current widespread mutation combination in China, and the emergence of high resistance to fluoroquinolones is driven by the ParC S80R mutation. Widespread resistance to fluoroquinolones poses a substantial challenge to the prevention and treatment of infections caused by Mycoplasma species; thus, increased vigilance and surveillance of M. bovis infections in cattle will be needed to prevent disease spread.
Dr. Lan is a postdoctoral research scientist at the Mycoplasma Research Group in Bacterial Disease Team, State Key Laboratory for Animal Disease Control and Prevention and Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences. Her research interests focus on the pathogenesis, prevention, and control of pathogenic Mycoplasma spp. in animals and M. pneumoniae in humans.
Acknowledgment
This work was supported by grants from the Key Program of the Natural Science Foundation of Gansu Province (grant no. 22JR5RA409), National Natural Science Foundation of China (grant no. 32373019), and Chinese Academy of Agricultural Science and Technology Innovation Project (grant no. CAAS-ASTIP-JBGS-20210701).
References
- Kinnear A, Waldner M, McAllister TA, Zaheer R, Register K, Jelinski M. Application of four genotyping methods to Mycoplasma bovis isolates derived from western Canadian feedlot cattle. J Clin Microbiol. 2021;59:
e0004421 . DOIPubMedGoogle Scholar - Nicholas RAJ, Ayling RD. Mycoplasma bovis: disease, diagnosis, and control. Res Vet Sci. 2003;74:105–12. DOIPubMedGoogle Scholar
- Hale HH, Helmboldt CF, Plastridge WN, Stula EF. Bovine mastitis caused by a Mycoplasma species. Cornell Vet. 1962;52:582–91.PubMedGoogle Scholar
- Qi J, Guo A, Cui P, Chen Y, Mustafa R, Ba X, et al. Comparative geno-plasticity analysis of Mycoplasma bovis HB0801 (Chinese isolate). PLoS One. 2012;7:
e38239 . DOIPubMedGoogle Scholar - Niu J, Li K, Pan H, Gao X, Li J, Wang D, et al. Epidemiological survey of Mycoplasma bovis in yaks on the Qinghai Tibetan Plateau, China. Biomed Res Int. 2021;2021:
6646664 . DOIPubMedGoogle Scholar - Guo Y, Luo H, Guo S, Lei Y, Li Y, He S. Multi-locus sequence typing of Mycoplasma bovis to assess its genetic diversity from 2009 to 2018 in Ningxia Hui Autonomous Region, China. BMC Vet Res. 2020;16:454. DOIPubMedGoogle Scholar
- Gautier-Bouchardon AV. Antimicrobial resistance in Mycoplasma spp. Microbiol Spectr. 2018;6:. DOIGoogle Scholar
- Lysnyansky I, Ayling RD. Mycoplasma bovis: mechanisms of resistance and trends in antimicrobial susceptibility. Front Microbiol. 2016;7:595. DOIPubMedGoogle Scholar
- Gautier-Bouchardon AV, Ferré S, Le Grand D, Paoli A, Gay E, Poumarat F. Overall decrease in the susceptibility of Mycoplasma bovis to antimicrobials over the past 30 years in France. PLoS One. 2014;9:
e87672 . DOIPubMedGoogle Scholar - Niu J, Yan M, Xu J, Xu Y, Chang Z, Sizhu S. The resistance mechanism of Mycoplasma bovis from yaks in Tibet to fluoroquinolones and aminoglycosides. Front Vet Sci. 2022;9:
840981 . DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: July 16, 2025
1These senior authors contributed equally to this article.
Table of Contents – Volume 31, Number 8—August 2025
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Shengli Chen and Yuefeng Chu, State Key Laboratory for Animal Disease Control and Prevention, College of Veterinary Medicine, Lanzhou University, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, No. 1, Xujiaping, Chengguan District, Lanzhou Gansu Province, 730046, China
Top