Volume 23, Number 4—April 2017
Dispatch
Antiviral Drug–Resistant Influenza B Viruses Carrying H134N Substitution in Neuraminidase, Laos, February 2016
Figure 1
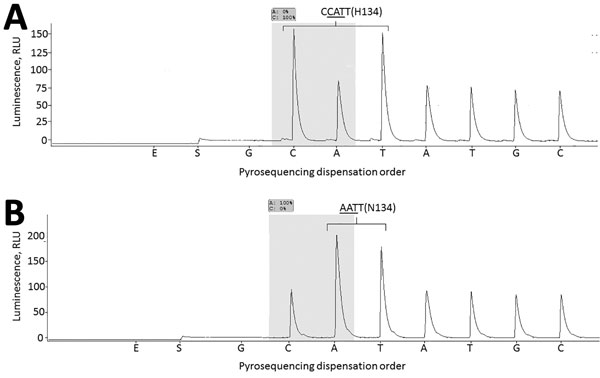
Figure 1. Neuraminidase gene segment (nts 399–497) of influenza B/Laos/0080/2016 virus carrying NA-H134 (A) and B/Laos/0654/2016, NA-N134 (B). RNA extracted from respiratory specimens was used for reverse transcription PCR (RT-PCR) amplification. Two primers, NA-B-242F (5′-CATACCCGCGTTTATCTTGC-3′, forward primer) and NA-B-426Rb (biotin-5′-CTGTCTCCTCTTGTTCCATTGTAG-3′, reverse biotinylated primer) were used in RT-PCR, essentially as described previously (10); primer NA-B-378Fs (5′-TGCAAACACTTTGCTTTAAC-3′) was used for pyrosequencing. Underlining indicates nucleotide triplet encoding amino acid residue 134.Shading indicates the nucleotides used to determine the proportion of H134 and N134 neuraminidase variants. Pyrosequencing dispensation order: E-Enzyme mixture; S-substrate mixture; G, C, A and T – nucleotides dGTP, dCTP; dATPαS and dTTP, correspondingly.
References
- Burnham AJ, Baranovich T, Govorkova EA. Neuraminidase inhibitors for influenza B virus infection: efficacy and resistance. Antiviral Res. 2013;100:520–34. DOIPubMedGoogle Scholar
- Budd A, Blanton L, Kniss K, Smith S, Mustaquim D, Davlin SL, et al. Update: influenza activity—United States and worldwide, May 22–September 10, 2016. MMWR Morb Mortal Wkly Rep. 2016;65:1008–14. DOIPubMedGoogle Scholar
- Davlin SL, Blanton L, Kniss K, Mustaquim D, Smith S, Kramer N, et al. Influenza activity—United States, 2015–16 season and composition of the 2016–17 influenza vaccine. MMWR Morb Mortal Wkly Rep. 2016;65:567–75. DOIPubMedGoogle Scholar
- Okomo-Adhiambo M, Mishin VP, Sleeman K, Saguar E, Guevara H, Reisdorf E, et al. Standardizing the influenza neuraminidase inhibition assay among United States public health laboratories conducting virological surveillance. Antiviral Res. 2016;128:28–35. DOIPubMedGoogle Scholar
- World Health Organization. Meetings of the WHO working group on surveillance of influenza antiviral susceptibility – Geneva, November 2011 and June 2012. Wkly Epidemiol Rec. 2012;87:369–74.PubMedGoogle Scholar
- Colman PM, Hoyne PA, Lawrence MC. Sequence and structure alignment of paramyxovirus hemagglutinin-neuraminidase with influenza virus neuraminidase. J Virol. 1993;67:2972–80.PubMedGoogle Scholar
- Kawai N, Ikematsu H, Iwaki N, Maeda T, Satoh I, Hirotsu N, et al. A comparison of the effectiveness of oseltamivir for the treatment of influenza A and influenza B: a Japanese multicenter study of the 2003-2004 and 2004-2005 influenza seasons. Clin Infect Dis. 2006;43:439–44. DOIPubMedGoogle Scholar
- Monto AS, McKimm-Breschkin JL, Macken C, Hampson AW, Hay A, Klimov A, et al. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob Agents Chemother. 2006;50:2395–402. DOIPubMedGoogle Scholar
- Sugaya N, Mitamura K, Yamazaki M, Tamura D, Ichikawa M, Kimura K, et al. Lower clinical effectiveness of oseltamivir against influenza B contrasted with influenza A infection in children. Clin Infect Dis. 2007;44:197–202. DOIPubMedGoogle Scholar
- Sheu TG, Deyde VM, Garten RJ, Klimov AI, Gubareva LV. Detection of antiviral resistance and genetic lineage markers in influenza B virus neuraminidase using pyrosequencing. Antiviral Res. 2010;85:354–60. DOIPubMedGoogle Scholar
- Takashita E, Meijer A, Lackenby A, Gubareva L, Rebelo-de-Andrade H, Besselaar T, et al. Global update on the susceptibility of human influenza viruses to neuraminidase inhibitors, 2013-2014. Antiviral Res. 2015;117:27–38. DOIPubMedGoogle Scholar
- Little K, Leang SK, Butler J, Baas C, Harrower B, Mosse J, et al. Zanamivir-resistant influenza viruses with Q136K or Q136R neuraminidase residue mutations can arise during MDCK cell culture creating challenges for antiviral susceptibility monitoring. Euro Surveill. 2015;20:30060. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Human influenza virus real-time RT-PCR detection and characterization panel. 510(k) summary. 2008. http://www.accessdata.fda.gov/cdrh_docs/pdf8/k080570.pdf.
- Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, et al. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J Virol. 2009;83:10309–13. DOIPubMedGoogle Scholar
- Ma LC, Guan R, Hamilton K, Aramini JM, Mao L, Wang S, et al. A second RNA-binding site in the NS1 protein of influenza B virus. Structure. 2016;24:1562–72. DOIPubMedGoogle Scholar