Volume 23, Number 4—April 2017
Dispatch
Characterization of Highly Pathogenic Avian Influenza Virus A(H5N6), Japan, November 2016
Figure 2
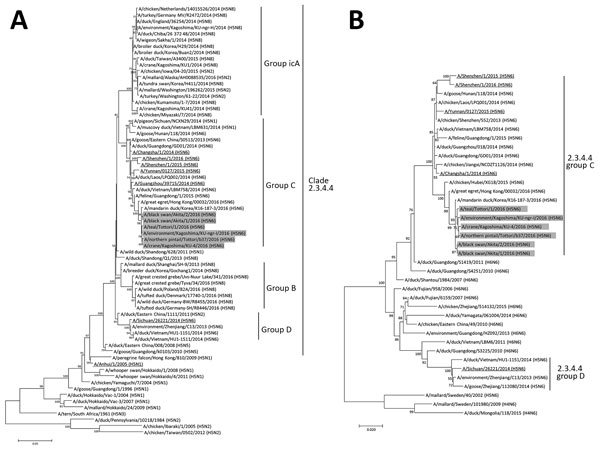
Figure 2. Phylogenetic trees of the HA and NA gene segments of highly pathogenic avian influenza virus A(H5N6) isolated in Japan. The nucleotide sequences of the H5 HA (A) and N6 NA (B) genes were analyzed by the maximum-likelihood method along with the corresponding genes of reference strains using MEGA 7.0 software (http://www.megasoftware.net/). Horizontal distances are proportional to the minimum number of nucleotide differences required to join nodes and sequences. Numbers at the nodes indicate the probability of confidence levels in a bootstrap analysis with 1,000 replications. Gray indicates viruses isolated in this study; underlining indicates viruses isolated in humans. The H5 HA gene sequences are classified into genetic clades as defined by Lee et al. (9). Scale bars indicate nucleotide substitutions per site. HA, hemagglutinin; NA, neuraminidase.
References
- Smith GJ, Donis RO. World Health Organization/World Organisation for Animal Health/Food and Agriculture Organization (WHO/OIE/FAO) H5 Evolution Working Group. Nomenclature updates resulting from the evolution of avian influenza A(H5) virus clades 2.1.3.2a, 2.2.1, and 2.3.4 during 2013–2014. Influenza Other Respi Viruses. 2015;9:271–6. DOIGoogle Scholar
- Claes F, Morzaria SP, Donis RO. Emergence and dissemination of clade 2.3.4.4 H5Nx influenza viruses-how is the Asian HPAI H5 lineage maintained. Curr Opin Virol. 2016;16:158–63. DOIPubMedGoogle Scholar
- Global Consortium for H5N8 and Related Influenza Viruses. Role for migratory wild birds in the global spread of avian influenza H5N8. Science. 2016;354:213–7. DOIPubMedGoogle Scholar
- Bi Y, Chen Q, Wang Q, Chen J, Jin T, Wong G, et al. Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe. 2016;20:810–21. DOIPubMedGoogle Scholar
- Ozawa M, Matsuu A, Tokorozaki K, Horie M, Masatani T, Nakagawa H, et al. Genetic diversity of highly pathogenic H5N8 avian influenza viruses at a single overwintering site of migratory birds in Japan, 2014/15. Euro Surveill. 2015;20:21132. DOIPubMedGoogle Scholar
- Usui T, Soda K, Tomioka Y, Ito H, Yabuta T, Takakuwa H, et al. Characterization of clade 2.3.4.4 H5N8 highly pathogenic avian influenza viruses from wild birds possessing atypical hemagglutinin polybasic cleavage sites. Virus Genes. 2016.PubMedGoogle Scholar
- Tanikawa T, Kanehira K, Tsunekuni R, Uchida Y, Takemae N, Saito T. Pathogenicity of H5N8 highly pathogenic avian influenza viruses isolated from a wild bird fecal specimen and a chicken in Japan in 2014. Microbiol Immunol. 2016;60:243–52. DOIPubMedGoogle Scholar
- World Organization for Animal Health. Update on highly pathogenic avian influenza in animals (type H5 and H7) [cited 2016 Dec 4]. http://www.oie.int/en/animal-health-in-the-world/update-on-avian-influenza/2016/
- Lee DH, Bahl J, Torchetti MK, Killian ML, Ip HS, DeLiberto TJ, et al. Highly pathogenic avian influenza viruses and generation of novel reassortants, United States, 2014–2015. Emerg Infect Dis. 2016;22:1283–5. DOIPubMedGoogle Scholar
- Lee DH, Sharshov K, Swayne DE, Kurskaya O, Sobolev I, Kabilov M, et al. Novel reassortant clade 2.3.4.4 avian influenza A(H5N8) virus in wild aquatic birds, Russia, 2016. Emerg Infect Dis. 2017;23:359–60. DOIPubMedGoogle Scholar
- Jiao P, Cao L, Yuan R, Wei L, Song Y, Shen D, et al. Complete genome sequence of an H10N8 avian influenza virus isolated from a live bird market in Southern China. J Virol. 2012;86:7716. DOIPubMedGoogle Scholar
- Guo H, de Vries E, McBride R, Dekkers J, Peng W, Bouwman KM, et al. Highly pathogenic influenza A(H5Nx) viruses with altered H5 receptor-binding specificity. Emerg Infect Dis. 2017;23:220–31. DOIPubMedGoogle Scholar
- Hiono T, Okamatsu M, Igarashi M, McBride R, de Vries RP, Peng W, et al. Amino acid residues at positions 222 and 227 of the hemagglutinin together with the neuraminidase determine binding of H5 avian influenza viruses to sialyl Lewis X. Arch Virol. 2016;161:307–16. DOIPubMedGoogle Scholar
- Hiono T, Ohkawara A, Ogasawara K, Okamatsu M, Tamura T, Chu DH, et al. Genetic and antigenic characterization of H5 and H7 influenza viruses isolated from migratory water birds in Hokkaido, Japan and Mongolia from 2010 to 2014. Virus Genes. 2015;51:57–68. DOIPubMedGoogle Scholar
- Yamamoto N, Sakoda Y, Motoshima M, Yoshino F, Soda K, Okamatsu M, et al. Characterization of a non-pathogenic H5N1 influenza virus isolated from a migratory duck flying from Siberia in Hokkaido, Japan, in October 2009. Virol J. 2011;8:65. DOIPubMedGoogle Scholar
1These authors contributed equally to this article.