Volume 23, Number 6—June 2017
Dispatch
Isolated Case of Marburg Virus Disease, Kampala, Uganda, 2014
Cite This Article
Citation for Media
Abstract
In September 2014, a single fatal case of Marburg virus was identified in a healthcare worker in Kampala, Uganda. The source of infection was not identified, and no secondary cases were identified. We describe the rapid identification, laboratory diagnosis, and case investigation of the third Marburg virus outbreak in Uganda.
Marburg virus disease (MVD) is caused by Marburg virus (MARV; family Filoviridae, which also includes Ebola viruses). The disease was first discovered in 1967 in Marburg and Frankfurt, Germany, after laboratory workers were infected from monkeys imported from Uganda (1). Thereafter, sporadic cases and outbreaks of MVD have been documented in South Africa, Kenya, the Democratic Republic of the Congo, Angola, Uganda, the Netherlands, and the United States (2). MVD remains of great public health importance because of the case-fatality rate, which can be as high as 90%, and documented human-to-human transmission, with associated socioeconomic consequences.
Uganda has experienced previous outbreaks of MVD, resulting in fatalities and socioeconomic effects from loss of tourism. The first recorded outbreak in Uganda occurred in the Kamwenge district in 2007, where 4 MVD cases were confirmed in miners at the Kitaka mine (3). A second, larger outbreak occurred in the western Uganda districts of Kabale, Ibanda, and Kamwenge in 2012 (4) and was also linked to mining activity in the Ibanda district. In addition, tourists from the United States and the Netherlands were infected with MARV in western Uganda when they visited Python Cave in Queen Elizabeth National Park in 2008 (5,6). Both Python Cave and the Kitaka mine are inhabited by Egyptian fruit bats (Rousettus aegyptiacus), the host reservoir of MARV (7).
In 2014, a fatal case of MVD occurred in Uganda. We report here on the field and laboratory investigation of this case, including possible sources of infection.
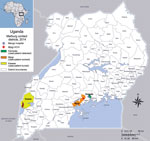
On September 23, 2014, a healthcare worker was admitted to Mengo Hospital in Kampala, Uganda, with a febrile illness suspected to be viral hemorrhagic fever (VHF). The patient, a 30-year-old man, was a radiographer who worked at Mengo Hospital and part-time at Mpigi Health Center IV in the Mpigi district, ≈30 km south of Kampala (Figure 1). His symptoms began on September 17, 2014. A rapid diagnostic test result was positive for malaria, and the patient was given intravenous ceftriaxone, 5% dextrose, and artesunate. However, his condition continued to deteriorate.
On September 26, he began to display hemorrhagic signs, notably profuse bleeding from body orifices. Clinical findings included fever (temperature 38.4°C), nausea, vomiting, diarrhea, musculoskeletal pain, abdominal pain, headache, sore throat, difficulty swallowing, difficulty breathing, anorexia, bleeding from the nose, bloody stools, vomiting blood, and upper gastrointestinal tract bleeding. He died on September 28.
Whole blood and serum samples collected on September 28 were sent to the Uganda Virus Research Institute (UVRI)/US Centers for Disease Control and Prevention (CDC) VHF laboratory in Entebbe for testing on September 30. We performed diagnostic testing by real-time reverse transcription PCR (RT-PCR) targeting the MARV NP gene, antigen detection, and MARV IgM ELISA on the suspected sample as described (8). We detected MARV by RT-PCR, but antigen detection and IgM serologic results were negative.
We obtained independent confirmation from duplicate whole blood and serum samples by using RT-PCR primers and probes targeting MARV VP40 gene and a commercial filovirus PCR screening assay (Altona Diagnostics, Hamburg, Germany) (9) (Table). On the basis of the positive RT-PCR results, we confirmed MVD in this patient. We shipped specimens to the CDC Viral Special Pathogens Branch, Division of High-Consequence Pathogens and Pathology, National Center for Emerging and Zoonotic Infectious Diseases (Atlanta, GA, USA), for further testing, including virus isolation and sequencing. A virus isolate (812601) was generated from the clinical specimen after a single passage in cell culture (Vero E6).
After laboratory confirmation, a multidisciplinary team from UVRI and CDC-Uganda performed the initial outbreak investigations at Mengo Hospital and Mpigi Health Center IV. The investigation team provided the outbreak case definition, and details of case contacts were obtained from both health facilities. An investigation was also performed in Kasese District, where the health worker was taken for burial. In addition, an ecologic investigation was conducted to identify roosting sites nearby for the potential presence of R. aegyptiacus bats.
We created an outbreak database by using the Epi Info Viral Hemorrhagic Fever outbreak management application (10). All case and contact data were managed by the Uganda Ministry of Health Public Health Emergency Operations Center. We identified 197 close contacts, who were followed for 21 days. During the course of follow-up, 33 (16.2%) of the 197 contacts converted to suspected case-patients by exhibiting clinically compatible signs or symptoms matching the outbreak case definition. Blood samples from suspected case-patients were tested at the UVRI/CDC laboratory in Entebbe; all were negative for MARV by RT-PCR and serologic analysis.
We describe the second single-case filovirus outbreak detected in Uganda; a case of infection with Sudan ebolavirus was reported in Luwero District in 2011 (11). The investigation was unable to identify a conclusive source of infection, including evidence of the natural reservoir host near where the infected patient was working or residing or potential cases in persons who visited health centers before this case was confirmed. No secondary cases arose from contact with the initial case-patient.
The patient tested positive for malaria but later tested positive for MARV by RT-PCR and serologic analysis. These findings indicate that co-infection of viral hemorrhagic fever (VHF) and other tropical infectious diseases can confound diagnosis and delay early detection, potentially resulting in large outbreaks that are much harder to control, as was seen during the 2014–2015 Ebola virus outbreak in West Africa (12). This finding emphasizes the need for continued surveillance and awareness even when other, more common pathogens are initially suspected.
The finding of no secondary cases in this investigation can be attributed, in part, to use of infection control practices and personal protective equipment when first encountering a suspected VHF case. Because Uganda has experienced 10 VHF outbreaks since 2011, increased awareness and use of personal protective equipment and infection control practices have greatly limited secondary transmission, especially in the healthcare setting. Routine use of gloves, protective gowns, and chlorine is now more common in lower-level healthcare facilities in Uganda; these protective products were used at both Mengo Hospital and Mpigi Health Center IV.
The full genomic sequence of this MARV isolate (812601; GenBank accession no. KP985768) falls into a cluster that consists mostly of MARV sequences isolated from bats. The closest related sequence was obtained from MARV isolated from a juvenile male R. aegyptiacus bat (Q843) captured in August 2009 in Python Cave, Queen Elizabeth National Park (Figure 2). Other viral sequences from bat specimens in this clade were from bats collected from either Python Cave or the Kitaka mine during 2007–2009. The most closely related human sequence (01Uga07) was from a miner who worked in the Kitaka mine in July 2007 (3,7). Because of the wide genetic diversity of MARVs, there is no definitive way to identify where this patient may have become infected.
We do not know why the patient did not transmit MARV to any of his close contacts. We can assume that none of the contacts had substantial exposure to the patient while he was infectious. The relatively small size of this and previous filovirus outbreaks in Uganda can be attributed to enhanced VHF surveillance, rapid case identification, laboratory testing, and investments from the global health security agenda in rapid sample transportation to the national VHF reference laboratory for diagnostic testing (13). VHF surveillance continues to be a top priority for Uganda, and the VHF surveillance program continues to play a crucial role in detecting, responding to, and helping to control these outbreaks.
Dr. Nyakarahuka is a zoonotic disease epidemiologist at the Uganda Virus Research Institute in Entebbe, Uganda, with a background in veterinary medicine and public health. His work specifically focuses on the surveillance, detection and investigation of viral hemorrhagic fevers, including Ebola and Marburg viruses, and he specializes in characterizing the natural ecology of hemorrhagic fever viruses.
Acknowledgment
We thank the staff at Mengo Hospital, Kampala, and Mpigi Health Center IV; the initial investigation team, which included members from UVRI and CDC-Uganda for field and laboratory investigation of the outbreak; the Ministry of Health National Task Force on Epidemic Preparedness and Response; World Health Organization; Kampala City Council Authority; Kasese and Mpigi local governments; CDC Uganda, CDC, and UVRI Motorpool for transportation; and Médecins Sans Frontières for establishing an isolation ward at Mulago Hospital. We especially thank UVRI for its support of the VHF surveillance program and diagnostics laboratory. We also thank the United States Agency for International Development, African Field Epidemiology Network, and the United Nations Children’s Fund, which all participated and contributed to the Marburg outbreak response and provided support to the Uganda Ministry of Health.
References
- Siegert R, Shu HL, Slenczka W. [Isolation and identification of the “Marburg virus”] [in German]. Dtsch Med Wochenschr. 1968;93:604–12. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Chronology of Marburg hemorrhagic fever outbreaks; known cases and outbreaks of Marburg hemorrhagic fever, in chronological order [cited 2014 Oct 29]. http://www.cdc.gov/vhf/marburg/resources/outbreak-table.html
- Adjemian J, Farnon EC, Tschioko F, Wamala JF, Byaruhanga E, Bwire GS, et al. Outbreak of Marburg hemorrhagic fever among miners in Kamwenge and Ibanda Districts, Uganda, 2007. J Infect Dis. 2011;204(Suppl 3):S796–9. DOIPubMedGoogle Scholar
- Knust B, Schafer IJ, Wamala J, Nyakarahuka L, Okot C, Shoemaker T, et al. Multidistrict outbreak of Marburg virus disease—Uganda, 2012. J Infect Dis. 2015;212(Suppl 2):S119–28. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention (CDC). Imported case of Marburg hemorrhagic fever - Colorado, 2008. MMWR Morb Mortal Wkly Rep. 2009;58:1377–81.PubMedGoogle Scholar
- Timen A, Koopmans MP, Vossen AC, van Doornum GJ, Günther S, van den Berkmortel F, et al. Response to imported case of Marburg hemorrhagic fever, the Netherland. Emerg Infect Dis. 2009;15:1171–5. DOIPubMedGoogle Scholar
- Towner JS, Amman BR, Sealy TK, Carroll SA, Comer JA, Kemp A, et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5:e1000536. DOIPubMedGoogle Scholar
- Ksiazek TG, West CP, Rollin PE, Jahrling PB, Peters CJ. ELISA for the detection of antibodies to Ebola viruses. J Infect Dis. 1999;179(Suppl 1):S192–8. DOIPubMedGoogle Scholar
- Diagnostics A. RealStar Filovirus Screen RT-PCR Kit 1.0, 2014 [cited 2016 Jun 9]. http://www.altona-diagnostics.com/tl_files/website/downloads/RealStar_Filovirus Screen RT-PCR Kit 10_CE_WEB_2014-08-29.pdf
- Schafer IJ, Knudsen E, McNamara LA, Agnihotri S, Rollin PE, Islam A. The Epi Info Viral Hemorrhagic Fever (VHF) application: a resource for outbreak data management and contact tracing in the 2014–2016 West Africa Ebola epidemic. J Infect Dis. 2016;214(suppl 3):S122–36. DOIPubMedGoogle Scholar
- Shoemaker T, MacNeil A, Balinandi S, Campbell S, Wamala JF, McMullan LK, et al. Reemerging Sudan Ebola virus disease in Uganda, 2011. Emerg Infect Dis. 2012;18:1480–3. DOIPubMedGoogle Scholar
- The Lancet. The Lancet. 1 year on—lessons from the Ebola outbreak for WHO. Lancet. 2015;385:1152. DOIGoogle Scholar
- Borchert JN, Tappero JW, Downing R, Shoemaker T, Behumbiize P, Aceng J, et al.; Centers for Disease Control and Prevention (CDC). Rapidly building global health security capacity—Uganda demonstration project, 2013. MMWR Morb Mortal Wkly Rep. 2014;63:73–6.PubMedGoogle Scholar
Figures
Table
Cite This ArticleTable of Contents – Volume 23, Number 6—June 2017
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Trevor R. Shoemaker, Centers for Disease Control and Prevention, P.O. Box 7007, Plot 1577 Ggaba Road, Kampala, Uganda
Top