Volume 23, Number 9—September 2017
Dispatch
Emergence of Plasmid-Mediated Fosfomycin-Resistance Genes among Escherichia coli Isolates, France
Abstract
FosA, a glutathione S-transferase that inactivates fosfomycin, has been reported as the cause of enzymatic resistance to fosfomycin. We show that multiple lineages of FosA-producing extended spectrum β-lactamase Escherichia coli have circulated in France since 2012, potentially reducing the efficacy of fosfomycin in treating infections with antimicrobial drug–resistant gram-negative bacilli.
Fosfomycin is a broad-spectrum bactericidal antibiotic commonly used in Europe as a first-line oral agent for uncomplicated urinary tract infection (1). In France, it is the only first-line antimicrobial drug recommended for treatment of cystitis (97% susceptibility) and is used in 20%–30% of such treatments (2). However, it is receiving renewed worldwide attention as one of the most active agents for sparing carbapenems in extended spectrum β-lactamase (ESBL)–producing isolates and for treatment of carbapeneme-resistant Enterobacteriaceae (CRE) in combination with colistin (3). In France, intravenous fosfomycin (3–4 g 4×/d) is used in combination with other drugs for the treatment of multidrug-resistant infections.
The evaluation of fosfomycin susceptibility in clinical strains is widely performed, but the molecular bases are rarely documented. Fosfomycin inhibits the initial step in peptidoglycan synthesis by irreversibly blocking MurA in both gram-positive and -negative bacteria. It is imported through the inner membrane through the glycerol-3-phosphate (G3P) transporter GlpT and the glucose-6-phosphate (G6P) transporter UhpT. Decreased expression or mutations in glpT or uhpT genes are the most frequent events leading to lowered susceptibility, whereas modification of the fosfomycin target MurA seems to be rare in clinical isolates (4). Another mechanism is the production of FosA, a glutathione S-transferase that inactivates fosfomycin by addition of a glutathione residue. This mechanism is particularly relevant because it is disseminative and frequently associated with ESBL-producing Escherichia coli. Since 2006, researchers in several countries in East Asia have described plasmid-mediated fosA3 and, less frequently, fosA5 (formerly fosKp96), which is mostly associated with CTX-M and co-harbored on a conjugative plasmid. Some studies have focused on human clinical strains in China (5), South Korea (6), or Japan (7), and others have addressed veterinary strains isolated throughout China from pets (8), livestock (9), or animal fodder (10). In 2016, Portugal reported the first imported case of a travel-related infection in Europe with an E. coli strain co-expressing fosA3 and CTX-M-15 (11). The possible dissemination of this gene is worrisome because fosA3 is generally surrounded by the IS26 insertion sequence on a composite transposon borne by the IncFII conjugative plasmid, which is known to be a dissemination vector of resistance genes worldwide. Here we report the prevalence and mechanisms of fosfomycin resistance among clinical human E. coli strains isolated in Paris, France.
We investigated the occurrence and molecular features of all fosfomycin-resistant E. coli isolated from hospitalized patients during a 12-month period (August 2014–July 2015). We performed bacterial identification by using VITEK 2 (BioMérieux, Marcy l’Etoile, France) and tested antibiotic susceptibility by using the disk diffusion method in accordance with 2016 Comité de l’Antibiogramme de la Société Française de Microbiologie/European Committee on Antimicrobial Susceptibility Testing guidelines (http://www.sfm-microbiologie.org/UserFiles/files/casfm/CASFM2016_V1_0_FEVRIER.pdf). We screened for fosfomycin resistance by using a 200-µg disk with a diameter cutoff of <13 mm. We determined MIC by using the Etest method with Muller-Hinton agar containing 25 mg/L G6P.
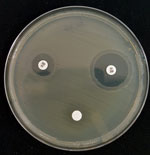
Among 1,354 E. coli isolates tested, 12 (0.9%) showed confirmed resistance (MIC >128 mg/L). We explored the mechanism of fosfomycin resistance by growing these isolates for 48 hours at 35°C in M9 minimal medium agar supplemented with either G3P or G6P at 0.2% as a sole carbon source. Lack of growth showed impaired fosfomycin transport (12). Of the 12 isolates, 7 were double auxotrophic mutants with G6P and G3P (mean MIC 384 mg/L), 3 were auxotrophic only for G3P (mean MIC 597 mg/L), and 2 were capable of using both substrates and exhibited high-level resistance (MIC >1,024 mg/L). Paradoxically, single and double auxotroph strains had lower mean MICs than the 2 nonauxotroph strains. Because transport deficit could not account for the observed phenotype, we screened by PCR and sequenced genes coding enzymatic glutathione S-transferase variants fosA, fosA2, fosA3, fosA4, fosA5, and fosC2 (Table 1). In parallel, we screened inhibition of glutathione S-transferase activity by using FosA inhibitor phosphonoformiate (Figure) as described by Nakamura et al. (12). Results of these 2 tests were in agreement, with each detecting an ESBL-producing strain with enzymatic activity encoded by the fosA3 gene (MIC >1,024 mg/L). Neither strain had been previously reported in France.
We determined the prevalence of enzymatic resistance to fosfomycin in ESBL-producing strains isolated since 2012 by using the same 2 tests. Surprisingly, among 23 strains resistant to fosfomycin with no epidemiologic link, 7 additional FosA3-producing and 1 FosA5-producing strains were detected, each with MIC >1,024 mg/L. Overall 83% of fosfomycin-resistant ESBL-producing E. coli with MIC >1,024 mg/L were FosA-positive. Auxotrophic tests showed that in addition of FosA production, fosfomycin transport was impaired in 6 strains. Chronologically, 2 strains were isolated in 2012, three in 2013, three in 2014, and two in 2015, meaning that FosA3-producing strains were present in France 6 years after the first isolation in Japan. The FosA5-producing strain was isolated from a clinical sample in France simultaneously with the original description of the strain in China (13). The sequencing of fosA5 showed 99% identity (with 96% coverage) with pHKU1, earlier described as an fosKP96-carrying IncN plasmid in Klebsiella pneumoniae (5).
Our sequencing of CTX-M genes showed that FosA3-producing strains were associated with CTX-M-15 (n = 5), CTX-M-55 (n = 3), and CTX-M-2 (n = 1), whereas the 1 FosA5-producing strain expressed CTX-M-14. We also conducted multilocus sequence typing and plasmid incompatibility group typing (14) (Table 2). These results show unambiguously that 8 strains of E. coli of different sequence types hosted 5 plasmid types that could be distinguished by their CTX-M variant and plasmid-incompatibility group types.
Nine out of 10 isolates yielded transconjugants in E. coli C600 (E. coli K12 derivative) or transformants in TOP10 (DH10B derivative) E. coli. All 9 of these isolates expressed high-level resistance to fosfomycin (MIC >1,024 mg/L), confirming that the observed resistance of the parent strain was indeed attributable to the fosA gene.
Although the prevalence of plasmid-mediated fosA3 genes in human clinical E. coli isolates has remained low in France since 2012, these genes are observed across numerous clones, sequence types, and molecular determinants and are always associated with ESBL CTX-M enzymes, suggesting multiple propagation events. Our results are consistent with FosA3-producing clinical strains previously isolated in Asia, which also co-express CTX-M enzymes. However, the CTX-M variant distribution between the strain in France and the strain in Asia are different, with CTX-M-15 having high prevalence in our collection. Medical records examination did not show a history of international travel in our patient population, and such a variety of fosfomycin-resistant E. coli lineages probably were not imported or transmitted. The broad use of oral fosfomycin has provided the opportunity to select for FosA producers. With the spread of CTX-M urinary tract infections in the community, the use of fosfomycin is likely to select for CTX-M–FosA co-producers and could lead to an increase of treatment failures with ESBL-producing organisms. Conversely, treatment of ESBL producers with fosfomycin should only be undertaken after testing for susceptibility because these ESBL producers can be linked to the same genetic determinant. Moreover, the indiscriminate use of the oral formulation in the community is jeopardizing the usefulness of this antimicrobial agent. While the world is bracing for an epidemic of infectious diseases bearing plasmid-mediated colistin resistance (15), a vast and ubiquitous reservoir for conjugative transmissible resistance to fosfomycin exists and can preclude its efficacy against extremely drug-resistant bacteria if the guidelines for the indiscriminate use of fosfomycin–trometamol are not urgently revised to safeguard this potent and well-tolerated agent. Because antimicrobial treatment of cystitis typically is motivated by concern for patient’s comfort, withholding treatment or the promotion of pivmecillinam as a first-line antimicrobial drug should seriously be considered.
Mr. Benzerara is an engineer and leads translational research, including emerging mechanisms of antimicrobial resistance, at the Hôpitaux Universitaires Est Parisiens Paris, France.
Acknowledgment
This work was funded by grants from the Assistance Publique Hôpitaux de Paris, France, and the Pierre et Marie Curie University, Paris, France.
References
- Société de Pathologie Infectieuse de Langue Française. Diagnostic et antibiothérapie des infections urinaires bactériennes communautaires de l’adulte [cited 2017 Jun 21]. http://www.infectiologie.com/UserFiles/File/spilf/recos/infections-urinaires-spilf.pdf
- Agence Nationale de Sécurité du Médicaments. Etude d’utilisation de la nitrofurantoïne en France—période mars 2012–février 2015 [cited 2017 Jun 21]. http://ansm.sante.fr/var/ansm_site/storage/original/application/d807c8e39321445201911cf314263f07.pdf
- Kaye KS, Gales AC, Dubourg G. Old antibiotics for multidrug-resistant pathogens: from in vitro activity to clinical outcomes. Int J Antimicrob Agents. 2017;49:542–8. DOIPubMedGoogle Scholar
- Castañeda-García A, Blázquez J, Rodríguez-Rojas A. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics (Basel). 2013;2:217–36. DOIPubMedGoogle Scholar
- Ho PL, Chan J, Lo WU, Lai EL, Cheung YY, Lau TC, et al. Prevalence and molecular epidemiology of plasmid-mediated fosfomycin resistance genes among blood and urinary Escherichia coli isolates. J Med Microbiol. 2013;62:1707–13. DOIPubMedGoogle Scholar
- Lee SY, Park YJ, Yu JK, Jung S, Kim Y, Jeong SH, et al. Prevalence of acquired fosfomycin resistance among extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J Antimicrob Chemother. 2012;67:2843–7. DOIPubMedGoogle Scholar
- Wachino J, Yamane K, Suzuki S, Kimura K, Arakawa Y. Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes. Antimicrob Agents Chemother. 2010;54:3061–4. DOIPubMedGoogle Scholar
- Hou J, Huang X, Deng Y, He L, Yang T, Zeng Z, et al. Dissemination of the fosfomycin resistance gene fosA3 with CTX-M β-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China. Antimicrob Agents Chemother. 2012;56:2135–8. DOIPubMedGoogle Scholar
- Chan J, Lo WU, Chow KH, Lai EL, Law PY, Ho PL. Clonal diversity of Escherichia coli isolates carrying plasmid-mediated fosfomycin resistance gene fosA3 from livestock and other animals. Antimicrob Agents Chemother. 2014;58:5638–9. DOIPubMedGoogle Scholar
- Hou J, Yang X, Zeng Z, Lv L, Yang T, Lin D, et al. Detection of the plasmid-encoded fosfomycin resistance gene fosA3 in Escherichia coli of food-animal origin. J Antimicrob Chemother. 2013;68:766–70. DOIPubMedGoogle Scholar
- Mendes AC, Rodrigues C, Pires J, Amorim J, Ramos MH, Novais Â, et al. Importation of fosfomycin resistance fosA3 gene to Europe. Emerg Infect Dis. 2016;22:346–8. DOIPubMedGoogle Scholar
- Nakamura G, Wachino J, Sato N, Kimura K, Yamada K, Jin W, et al. Practical agar-based disk potentiation test for detection of fosfomycin-nonsusceptible Escherichia coli clinical isolates producing glutathione S-transferases. J Clin Microbiol. 2014;52:3175–9. DOIPubMedGoogle Scholar
- Ma Y, Xu X, Guo Q, Wang P, Wang W, Wang M. Characterization of fosA5, a new plasmid-mediated fosfomycin resistance gene in Escherichia coli. Lett Appl Microbiol. 2015;60:259–64. DOIPubMedGoogle Scholar
- Compain F, Poisson A, Le Hello S, Branger C, Weill FX, Arlet G, et al. Targeting relaxase genes for classification of the predominant plasmids in Enterobacteriaceae. Int J Med Microbiol. 2014;304:236–42. DOIPubMedGoogle Scholar
- Al-Tawfiq JA, Laxminarayan R, Mendelson M. How should we respond to the emergence of plasmid-mediated colistin resistance in humans and animals? Int J Infect Dis. 2017;54:77–84. DOIPubMedGoogle Scholar
Figure
Tables
Cite This ArticleTable of Contents – Volume 23, Number 9—September 2017
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Guillaume Arlet, Hôpital Tenon – Bacteriology, 4 Rue de la Chine, Paris 75020 France
Top