Volume 24, Number 12—December 2018
Research
Genomic Characterization of β-Glucuronidase–Positive Escherichia coli O157:H7 Producing Stx2a
Figure 2
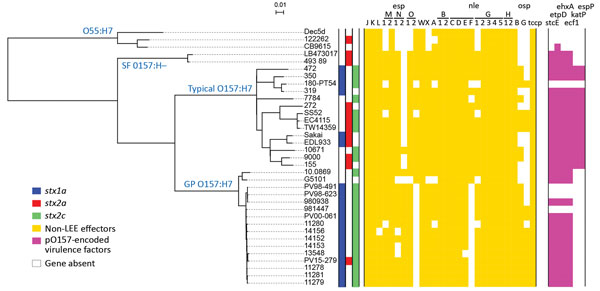
Figure 2. Whole-genome sequence-based phylogenetic analysis and repertoires of T3SS effectors and plasmid-encoding virulence factors from the study of Stx–producing E. coli O157:H7. The genome sequences of all the strains used in this study were aligned with the complete chromosome sequence of Sakai, and the single-nucleotide polymorphisms located in the 4,074,209-bp backbone sequence that were conserved in all the test strains were identified. After removing the recombinogenic single-nucleotide polymorphisms sites, we performed a concatenated alignment of 5,803 informative sites to generate the maximum-likelihood phylogeny. The conservation of T3SS effectors and plasmid-encoding virulence factors is shown in the tree. Colored boxes indicate the presence, and open boxes the absence, of each gene. GP, β-glucuronidase–positive; LEE, locus of enterocyte effacement; SF, sorbitol-fermenting. Scale bars indicate nucleotide substitutions per site.