Volume 24, Number 4—April 2018
Research
Phenotypic and Genotypic Characterization of Enterobacteriaceae Producing Oxacillinase-48–Like Carbapenemases, United States
Figure 2
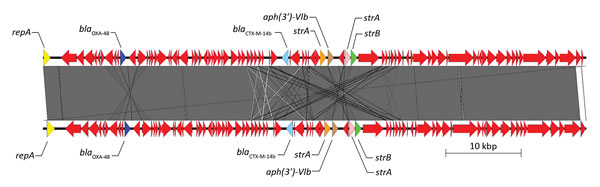
Figure 2. Sequence structure of 2 β-lactamase OXA-48 (blaOXA-48) plasmids tested during phenotypic and genotypic characterization of Enterobacteriaceae producing OXA-48–like carbapenemases, United States. Top plasmid is from isolate 23 in this study (pIncL_M_DHQP1400954) (72,093 bp), and bottom plasmid is from Chen et al. (44) (GenBank accession no. KP025948). Arrows indicate direction of transcription. Red arrows indicate other genes. Gray area indicates regions of homology, white lines indicate nonhomologous regions, and gray lines indicate inversions. aph, aminoglycoside; OXA, oxacillinase; repA, IncL/M type replicase; str, streptomycin.
References
- Gupta N, Limbago BM, Patel JB, Kallen AJ. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011;53:60–7. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention (CDC). Vital signs: carbapenem-resistant Enterobacteriaceae. MMWR Morb Mortal Wkly Rep. 2013;62:165–70.PubMedGoogle Scholar
- Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008;29:1099–106. DOIPubMedGoogle Scholar
- Walther-Rasmussen J, Høiby N. Class A carbapenemases. J Antimicrob Chemother. 2007;60:470–82. DOIPubMedGoogle Scholar
- Guh AY, Bulens SN, Mu Y, Jacob JT, Reno J, Scott J, et al. Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012–2013. JAMA. 2015;314:1479–87. DOIPubMedGoogle Scholar
- Lascols C, Peirano G, Hackel M, Laupland KB, Pitout JD. Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob Agents Chemother. 2013;57:130–6. DOIPubMedGoogle Scholar
- Mathers AJ, Hazen KC, Carroll J, Yeh AJ, Cox HL, Bonomo RA, et al. First clinical cases of OXA-48-producing carbapenem-resistant Klebsiella pneumoniae in the United States: the “menace” arrives in the new world. J Clin Microbiol. 2013;51:680–3. DOIPubMedGoogle Scholar
- Doi Y, O’Hara JA, Lando JF, Querry AM, Townsend BM, Pasculle AW, et al. Co-production of NDM-1 and OXA-232 by Klebsiella pneumoniae. Emerg Infect Dis. 2014;20:163–5. DOIPubMedGoogle Scholar
- Yang S, Hemarajata P, Hindler J, Li F, Adisetiyo H, Aldrovandi G, et al. Evolution and transmission of carbapenem-resistant Klebsiella pneumoniae expressing the blaOXA-232 gene during an institutional outbreak associated with endoscopic retrograde cholangiopancreatography. Clin Infect Dis. 2017;64:894–901. DOIPubMedGoogle Scholar
- Rojas LJ, Hujer AM, Rudin SD, Wright MS, Domitrovic TN, Marshall SH, et al. NDM-5 and OXA-181 beta-lactamases, a significant threat continues to spread in the Americas. Antimicrob Agents Chemother. 2017;61:e00454–17. DOIPubMedGoogle Scholar
- Poirel L, Héritier C, Tolün V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:15–22. DOIPubMedGoogle Scholar
- Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother. 2012;67:1597–606. DOIPubMedGoogle Scholar
- Oteo J, Hernández JM, Espasa M, Fleites A, Sáez D, Bautista V, et al. Emergence of OXA-48-producing Klebsiella pneumoniae and the novel carbapenemases OXA-244 and OXA-245 in Spain. J Antimicrob Chemother. 2013;68:317–21. DOIPubMedGoogle Scholar
- Sampaio JL, Ribeiro VB, Campos JC, Rozales FP, Magagnin CM, Falci DR, et al. Detection of OXA-370, an OXA-48-related class D β-lactamase, in Enterobacter hormaechei from Brazil. Antimicrob Agents Chemother. 2014;58:3566–7. DOIPubMedGoogle Scholar
- Meunier D, Vickers A, Pike R, Hill RL, Woodford N, Hopkins KL. Evaluation of the K-SeT R.E.S.I.S.T. immunochromatographic assay for the rapid detection of KPC and OXA-48-like carbapenemases. J Antimicrob Chemother. 2016;71:2357–9. DOIPubMedGoogle Scholar
- Pasteran F, Denorme L, Ote I, Gomez S, De Belder D, Glupczynski Y, et al. Rapid identification of OXA-48 and OXA-163 subfamilies in carbapenem-resistant gram-negative bacilli with a novel immunochromatographic lateral flow assay. J Clin Microbiol. 2016;54:2832–6. DOIPubMedGoogle Scholar
- Dortet L, Naas T. Noncarbapenemase OXA-48 variants (OXA-163 and OXA-405) falsely detected as carbapenemases by the β Carba test. J Clin Microbiol. 2017;55:654–5. DOIPubMedGoogle Scholar
- Gomez S, Pasteran F, Faccone D, Bettiol M, Veliz O, De Belder D, et al. Intrapatient emergence of OXA-247: a novel carbapenemase found in a patient previously infected with OXA-163-producing Klebsiella pneumoniae. Clin Microbiol Infect. 2013;19:E233–5. DOIPubMedGoogle Scholar
- Lyman M, Walters M, Lonsway D, Rasheed K, Limbago B, Kallen A. Notes from the field: carbapenem-resistant Enterobacteriaceae producing OXA-48-like carbapenemases—United States, 2010–2015. MMWR Morb Mortal Wkly Rep. 2015;64:1315–6. DOIPubMedGoogle Scholar
- Lascols C, Bonaparte S, Lonsway D, Johnson K, Robinson G, Rasheed K, et al. Snapshot of beta-lactam resistance in Enterobacteriaceae in the United States. Poster 1051. In: Abstracts of the 25th European Congress of Clinical Microbiology and Infectious Diseases, Copenhagen, Denmark, April 25–28, 2015. Abstract 1051 [cited 2018 Jan 26] https://www.escmid.org/dates_events/calendar/calendar_event/cal/2015/04/25/event/tx_cal_phpicalendar/25th_European_Congress_of_Clinical_Microbriology_and_Infectious_Diseases_ECCMID_2015/?tx_cal_controller%5Blastview%5D=view-search_event%7Cpage_id-130&cHash=fc21e6af2b4d39892216b2e8d30f0936
- Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard. 10th ed. (M07-A10). Wayne (PA): The Institute; 2015.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: twenty-seventh informational supplement. (M100-S27). Wayne (PA): The Institute; 2017.
- Conrad S, Oethinger M, Kaifel K, Klotz G, Marre R, Kern WV. gyrA mutations in high-level fluoroquinolone-resistant clinical isolates of Escherichia coli. J Antimicrob Chemother. 1996;38:443–55. DOIPubMedGoogle Scholar
- Kitchel B, Zhu W, Travis T, Limbago BM, Rasheed JK. Detection and evaluation of OXA-48 like carbapenemases by real-time PCR. Poster D-1139. In: Abstracts of the 53rd Interscience Conference on Antimicrobial Agents and Chemotherapy, Denver, Colorado, September 10–13, 2013 cited 2018 Jan 26]. https://www.medscape.com/viewcollection/32893
- Cox MP, Peterson DA, Biggs PJ, Solexa QA. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics. 2010;11:485. DOIPubMedGoogle Scholar
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. DOIPubMedGoogle Scholar
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. DOIPubMedGoogle Scholar
- Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–9. DOIPubMedGoogle Scholar
- Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. DOIPubMedGoogle Scholar
- de Man TJ, Limbago BM. SSTAR: a stand-alone easy-to-use antimicrobial resistance gene predictor. mSphere. 2016;1:pii: e00050-25. PMID: 27303709.DOIGoogle Scholar
- Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58:212–20. DOIPubMedGoogle Scholar
- Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–4. DOIPubMedGoogle Scholar
- Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–903. DOIPubMedGoogle Scholar
- Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–6. DOIPubMedGoogle Scholar
- Potron A, Nordmann P, Rondinaud E, Jaureguy F, Poirel L. A mosaic transposon encoding OXA-48 and CTX-M-15: towards pan-resistance. J Antimicrob Chemother. 2013;68:476–7. DOIPubMedGoogle Scholar
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. DOIPubMedGoogle Scholar
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. DOIPubMedGoogle Scholar
- Durfee T, Nelson R, Baldwin S, Plunkett G III, Burland V, Mau B, et al. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J Bacteriol. 2008;190:2597–606. DOIPubMedGoogle Scholar
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. DOIPubMedGoogle Scholar
- Karim A, Poirel L, Nagarajan S, Nordmann P. Plasmid-mediated extended-spectrum beta-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol Lett. 2001;201:237–41.PubMedGoogle Scholar
- McGann P, Snesrud E, Ong AC, Appalla L, Koren M, Kwak YI, et al. War wound treatment complications due to transfer of an IncN plasmid harboring bla(OXA-181) from Morganella morganii to CTX-M-27-producing sequence type 131 Escherichia coli. Antimicrob Agents Chemother. 2015;59:3556–62. DOIPubMedGoogle Scholar
- Potron A, Nordmann P, Lafeuille E, Al Maskari Z, Al Rashdi F, Poirel L. Characterization of OXA-181, a carbapenem-hydrolyzing class D beta-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2011;55:4896–9. DOIPubMedGoogle Scholar
- Potron A, Rondinaud E, Poirel L, Belmonte O, Boyer S, Camiade S, et al. Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class D β-lactamase from Enterobacteriaceae. Int J Antimicrob Agents. 2013;41:325–9. DOIPubMedGoogle Scholar
- Chen L, Al Laham N, Chavda KD, Mediavilla JR, Jacobs MR, Bonomo RA, et al. First report of an OXA-48-producing multidrug-resistant Proteus mirabilis strain from Gaza, Palestine. Antimicrob Agents Chemother. 2015;59:4305–7. DOIPubMedGoogle Scholar
- Papagiannitsis CC, Študentová V, Izdebski R, Oikonomou O, Pfeifer Y, Petinaki E, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry meropenem hydrolysis assay with NH4HCO3, a reliable tool for direct detection of carbapenemase activity. J Clin Microbiol. 2015;53:1731–5. DOIPubMedGoogle Scholar
- Tijet N, Boyd D, Patel SN, Mulvey MR, Melano RG. Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57:4578–80. DOIPubMedGoogle Scholar
- Chea N, Bulens SN, Kongphet-Tran T, Lynfield R, Shaw KM, Vagnone PS, et al. Improved phenotype-based definition for identifying carbapenemase producers among carbapenem-resistant Enterobacteriaceae. Emerg Infect Dis. 2015;21:1611–6. DOIPubMedGoogle Scholar
- Carrër A, Poirel L, Eraksoy H, Cagatay AA, Badur S, Nordmann P. Spread of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Istanbul, Turkey. Antimicrob Agents Chemother. 2008;52:2950–4. DOIPubMedGoogle Scholar
- Poirel L, Bonnin RA, Nordmann P. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother. 2012;56:559–62. DOIPubMedGoogle Scholar
- Villa L, Carattoli A, Nordmann P, Carta C, Poirel L. Complete sequence of the IncT-type plasmid pT-OXA-181 carrying the blaOXA-181 carbapenemase gene from Citrobacter freundii. Antimicrob Agents Chemother. 2013;57:1965–7. DOIPubMedGoogle Scholar
Page created: July 31, 2018
Page updated: July 31, 2018
Page reviewed: July 31, 2018
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.