Novel Enterobacter Lineage as Leading Cause of Nosocomial Outbreak Involving Carbapenemase-Producing Strains
Racha Beyrouthy, Marion Barets, Elodie Marion, Cédric Dananché, Olivier Dauwalder, Frédéric Robin, Lauraine Gauthier, Agnès Jousset, Laurent Dortet, François Guérin, Thomas Bénet, Pierre Cassier, Philippe Vanhems
1, and Richard Bonnet
1
Author affiliations: Centre Hospitalier Universitaire, Clermont-Ferrand, France (R. Beyrouthy, F. Robin, R. Bonnet); Centre National de Référence de la Résistance aux Antibiotiques, Clermont-Ferrand (R. Beyrouthy, F. Robin, R. Bonnet); Université Clermont Auvergne, Clermont-Ferrand (R. Beyrouthy, F. Robin, R. Bonnet); Institut National de la Santé et de la Recherche Médicale, Clermont-Ferrand (R. Beyrouthy, F. Robin, R. Bonnet); Institut National de la Recherche Agronomique, Clermont-Ferrand (R. Beyrouthy, F. Robin, R. Bonnet); Hôpital Édouard Herriot, Hospices Civils de Lyon, Lyon, France (M. Barets, E. Marion, C. Dananché, T. Bénet, P. Cassier, P. Vanhems); Groupement Hospitalier Est, Hospices Civils de Lyon, Lyon (O. Dauwalder); Centre International de Recherche en Infectiologie, Lyon (O. Dauwalder, T. Bénet, P. Vanhems),; Centre Hospitalier Universitaire Bicêtre, Paris, France (L. Gauthier, A. Jousset, L. Dortet); Centre National de Référence Associé de la Résistance aux Antibiotiques, Paris (L. Gauthier, A. Jousset, L. Dortet); Centre Hospitalier Universitaire, Caen, France (F. Guérin)
Main Article
Figure 7
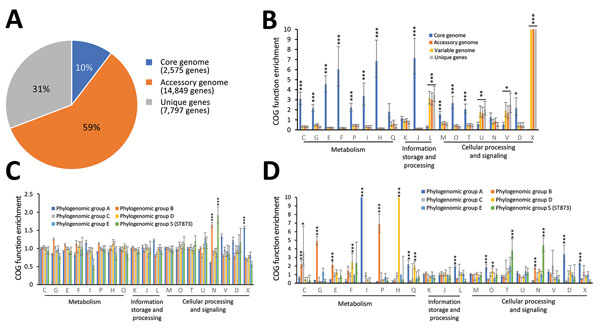
Figure 7. Pangenome analysis of metacluster Enterobacter hormaechei in study of nosocomial outbreak involving carbapenamase-producing Enterobacter strains, Lyon, France, January 12, 2014–December 31, 2015. A) Distribution of COGs); B) functional annotations in the pangenome; C) functional annotations in the variable genome (accessory genome + unique genes); and D) functional annotations for specific genes. Bar charts show the enrichment of COG categories as odds ratios; error bars indicate 95% CIs. Asterisks indicate certain COG categories that are significantly enriched: *p<0.05; **p<0.01;***p<0.001, all by Fisher exact test. Each COG category is identified by a 1-letter abbreviation: C, energy production and conversion; D, cell cycle control and mitosis; E, amino acid metabolism and transport; F, nucleotide metabolism and transport; G, carbohydrate metabolism and transport; H, coenzyme metabolism; I, lipid metabolism; J, translation; K, transcription; L, replication, recombination and repair; M, cell wall/membrane/envelope biogenesis; N, cell motility; O, post-translational modification, protein turnover, and chaperone functions; P, inorganic ion transport and metabolism; Q, secondary metabolism; T, signal transduction; U, intracellular trafficking and secretion; V, defense mechanisms; and X, mobilome. COG, clusters of orthologous groups.
Main Article
Page created: July 18, 2018
Page updated: July 18, 2018
Page reviewed: July 18, 2018
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
